Genome-Wide Identification and Characterization of the CsFHY3/FAR1 Gene Family and Expression Analysis under Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis)
- PMID: 33802900
- PMCID: PMC8002597
- DOI: 10.3390/plants10030570
Genome-Wide Identification and Characterization of the CsFHY3/FAR1 Gene Family and Expression Analysis under Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis)
Abstract
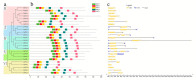
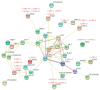
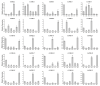
The FHY3/FAR1 transcription factor family, derived from transposases, plays important roles in light signal transduction, and in the growth and development of plants. However, the homologous genes in tea plants have not been studied. In this study, 25 CsFHY3/FAR1 genes were identified in the tea plant genome through a genome-wide study, and were classified into five subgroups based on their phylogenic relationships. Their potential regulatory roles in light signal transduction and photomorphogenesis, plant growth and development, and hormone responses were verified by the existence of the corresponding cis-acting elements. The transcriptome data showed that these genes could respond to salt stress and shading treatment. An expression analysis revealed that, in different tissues, especially in leaves, CsFHY3/FAR1s were strongly expressed, and most of these genes were positively expressed under salt stress (NaCl), and negatively expressed under low temperature (4 °C) stress. In addition, a potential interaction network demonstrated that PHYA, PHYC, PHYE, LHY, FHL, HY5, and other FRSs were directly or indirectly associated with CsFHY3/FAR1 members. These results will provide the foundation for functional studies of the CsFHY3/FAR1 family, and will contribute to the breeding of tea varieties with high light efficiency and strong stress resistance.
Keywords: CsFHY3/FAR1 family; biotic and abiotic stress; expression pattern; interaction network; light.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

