Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions
- PMID: 33802945
- PMCID: PMC8002816
- DOI: 10.3390/v13030497
Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions
Abstract
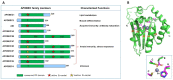
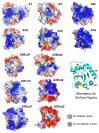
Apolipoprotein B mRNA editing catalytic polypeptide-like (APOBEC) proteins belong to a family of deaminase proteins that can catalyze the deamination of cytosine to uracil on single-stranded DNA or/and RNA. APOBEC proteins are involved in diverse biological functions, including adaptive and innate immunity, which are critical for restricting viral infection and endogenous retroelements. Dysregulation of their functions can cause undesired genomic mutations and RNA modification, leading to various associated diseases, such as hyper-IgM syndrome and cancer. This review focuses on the structural and biochemical data on the multimerization status of individual APOBECs and the associated functional implications. Many APOBECs form various multimeric complexes, and multimerization is an important way to regulate functions for some of these proteins at several levels, such as deaminase activity, protein stability, subcellular localization, protein storage and activation, virion packaging, and antiviral activity. The multimerization of some APOBECs is more complicated than others, due to the associated complex RNA binding modes.
Keywords: innate and acquired immunity; multimerization or oligomerization; mutation and cancer; structure; viral restriction.
Conflict of interest statement
The author declares no conflict of interest.
Figures





Similar articles
-
DNA Editing by APOBECs: A Genomic Preserver and Transformer.Trends Genet. 2016 Jan;32(1):16-28. doi: 10.1016/j.tig.2015.10.005. Epub 2015 Nov 20. Trends Genet. 2016. PMID: 26608778 Review.
-
Family-Wide Comparative Analysis of Cytidine and Methylcytidine Deamination by Eleven Human APOBEC Proteins.J Mol Biol. 2017 Jun 16;429(12):1787-1799. doi: 10.1016/j.jmb.2017.04.021. Epub 2017 May 4. J Mol Biol. 2017. PMID: 28479091 Free PMC article.
-
APOBECs orchestrate genomic and epigenomic editing across health and disease.Trends Genet. 2021 Nov;37(11):1028-1043. doi: 10.1016/j.tig.2021.07.003. Epub 2021 Aug 2. Trends Genet. 2021. PMID: 34353635 Review.
-
Modeling the Embrace of a Mutator: APOBEC Selection of Nucleic Acid Ligands.Trends Biochem Sci. 2018 Aug;43(8):606-622. doi: 10.1016/j.tibs.2018.04.013. Epub 2018 May 23. Trends Biochem Sci. 2018. PMID: 29803538 Free PMC article. Review.
-
The APOBEC Protein Family: United by Structure, Divergent in Function.Trends Biochem Sci. 2016 Jul;41(7):578-594. doi: 10.1016/j.tibs.2016.05.001. Epub 2016 Jun 6. Trends Biochem Sci. 2016. PMID: 27283515 Free PMC article. Review.
Cited by
-
APOBEC-1 cofactors regulate APOBEC3-induced mutations in hepatitis B virus.J Virol. 2025 Feb 25;99(2):e0187924. doi: 10.1128/jvi.01879-24. Epub 2025 Jan 27. J Virol. 2025. PMID: 39868801 Free PMC article.
-
Engineered deaminases as a key component of DNA and RNA editing tools.Mol Ther Nucleic Acids. 2023 Oct 20;34:102062. doi: 10.1016/j.omtn.2023.102062. eCollection 2023 Dec 12. Mol Ther Nucleic Acids. 2023. PMID: 38028200 Free PMC article. Review.
-
Human APOBEC3 Variations and Viral Infection.Viruses. 2021 Jul 14;13(7):1366. doi: 10.3390/v13071366. Viruses. 2021. PMID: 34372572 Free PMC article. Review.
-
Special Issue "APOBECs and Virus Restriction".Viruses. 2021 Aug 15;13(8):1613. doi: 10.3390/v13081613. Viruses. 2021. PMID: 34452478 Free PMC article.
-
The Roles of APOBEC-mediated RNA Editing in SARS-CoV-2 Mutations, Replication and Fitness.bioRxiv [Preprint]. 2022 Apr 7:2021.12.18.473309. doi: 10.1101/2021.12.18.473309. bioRxiv. 2022. Update in: Sci Rep. 2022 Sep 13;12(1):14972. doi: 10.1038/s41598-022-19067-x. PMID: 34981048 Free PMC article. Updated. Preprint.
References
-
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific Expression of Activation-induced Cytidine Deaminase (AID), a Novel Member of the RNA-editing Deaminase Family in Germinal Center B Cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

