Genetic Markers of Genome Rearrangements in Helicobacter pylori
- PMID: 33802974
- PMCID: PMC8002640
- DOI: 10.3390/microorganisms9030621
Genetic Markers of Genome Rearrangements in Helicobacter pylori
Abstract
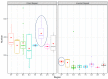
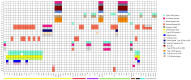
Helicobacter pylori exhibits a diverse genomic structure with high mutation and recombination rates. Various genetic elements function as drivers of this genomic diversity including genome rearrangements. Identifying the association of these elements with rearrangements can pave the way to understand its genome evolution. We analyzed the order of orthologous genes among 72 publicly available complete genomes to identify large genome rearrangements, and rearrangement breakpoints were compared with the positions of insertion sequences, genomic islands, and restriction modification genes. Comparison of the shared inversions revealed the conserved genomic elements across strains from different geographical locations. Some were region-specific and others were global, indicating that highly shared rearrangements and their markers were more ancestral than strain-or region-specific ones. The locations of genomic islands were an important factor for the occurrence of the rearrangements. Comparative genomics helps to evaluate the conservation of various elements contributing to the diversity across genomes.
Keywords: Helicobacter pylori; insertion sequences; inversion breakpoints; repeats.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

