viromeBrowser: A Shiny App for Browsing Virome Sequencing Analysis Results
- PMID: 33803225
- PMCID: PMC7999463
- DOI: 10.3390/v13030437
viromeBrowser: A Shiny App for Browsing Virome Sequencing Analysis Results
Abstract
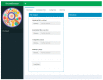
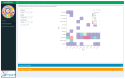
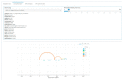
Experiments in which complex virome sequencing data is generated remain difficult to explore and unpack for scientists without a background in data science. The processing of raw sequencing data by high throughput sequencing workflows usually results in contigs in FASTA format coupled to an annotation file linking the contigs to a reference sequence or taxonomic identifier. The next step is to compare the virome of different samples based on the metadata of the experimental setup and extract sequences of interest that can be used in subsequent analyses. The viromeBrowser is an application written in the opensource R shiny framework that was developed in collaboration with end-users and is focused on three common data analysis steps. First, the application allows interactive filtering of annotations by default or custom quality thresholds. Next, multiple samples can be visualized to facilitate comparison of contig annotations based on sample specific metadata values. Last, the application makes it easy for users to extract sequences of interest in FASTA format. With the interactive features in the viromeBrowser we aim to enable scientists without a data science background to compare and extract annotation data and sequences from virome sequencing analysis results.
Keywords: NGS analysis; bioinformatics; data analysis; metagenomics; shiny; virome; visualization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function.Methods Mol Biol. 2016;1399:207-33. doi: 10.1007/978-1-4939-3369-3_13. Methods Mol Biol. 2016. PMID: 26791506
-
Hecatomb: an integrated software platform for viral metagenomics.Gigascience. 2024 Jan 2;13:giae020. doi: 10.1093/gigascience/giae020. Gigascience. 2024. PMID: 38832467 Free PMC article.
-
Entourage: all-in-one sequence analysis software for genome assembly, virus detection, virus discovery, and intrasample variation profiling.BMC Bioinformatics. 2024 Jun 24;25(1):222. doi: 10.1186/s12859-024-05846-y. BMC Bioinformatics. 2024. PMID: 38914932 Free PMC article.
-
Bioinformatics for NGS-based metagenomics and the application to biogas research.J Biotechnol. 2017 Nov 10;261:10-23. doi: 10.1016/j.jbiotec.2017.08.012. Epub 2017 Aug 18. J Biotechnol. 2017. PMID: 28823476 Review.
-
Evolution of selective-sequencing approaches for virus discovery and virome analysis.Virus Res. 2017 Jul 15;239:172-179. doi: 10.1016/j.virusres.2017.06.005. Epub 2017 Jun 3. Virus Res. 2017. PMID: 28583442 Free PMC article. Review.
References
-
- Xu B., Xu W., Li J., Dai L., Xiong C., Tang X., Yang Y., Mu Y., Zhou J., Ding J., et al. Metagenomic analysis of the Rhinopithecus bieti fecal microbiome reveals a broad diversity of bacterial and glycoside hydrolase profiles related to lignocellulose degradation. BMC Genom. 2015;16:174. doi: 10.1186/s12864-015-1378-7. - DOI - PMC - PubMed
-
- Naccache S.N., Federman S., Veeraraghavan N., Zaharia M., Lee D., Samayoa E., Bouquet J., Greninger A.L., Luk K.C., Enge B., et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–1192. doi: 10.1101/gr.171934.113. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

