The Role of circRNAs in Human Papillomavirus (HPV)-Associated Cancers
- PMID: 33803232
- PMCID: PMC7963196
- DOI: 10.3390/cancers13051173
The Role of circRNAs in Human Papillomavirus (HPV)-Associated Cancers
Abstract
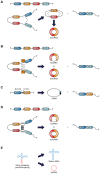
Circular RNAs (circRNAs) are a new class of "non-coding RNAs" that originate from non-sequential back-splicing of exons and/or introns of precursor messenger RNAs (pre-mRNAs). These molecules are generally produced at low levels in a cell-type-specific manner in mammalian tissues, but due to their circular conformation they are unaffected by the cell mRNA decay machinery. circRNAs can sponge multiple microRNAs or RNA-binding proteins and play a crucial role in the regulation of gene expression and protein translation. Many circRNAs have been shown to be aberrantly expressed in several cancer types, and to sustain specific oncogenic processes. Particularly, in virus-associated malignancies such as human papillomavirus (HPV)-associated anogenital carcinoma and oropharyngeal and oral cancers, circRNAs have been shown to be involved in tumorigenesis and cancer progression, as well as in drug resistance, and some are useful diagnostic and prognostic markers. HPV-derived circRNAs, encompassing the HPV E7 oncogene, have been shown to be expressed and to serve as transcript for synthesis of the E7 oncoprotein, thus reinforcing the virus oncogenic activity in HPV-associated cancers. In this review, we summarize research advances in the biogenesis of cell and viral circRNAs, their features and functions in the pathophysiology of HPV-associated tumors, and their importance as diagnostic, prognostic, and therapeutic targets in anogenital and oropharyngeal and oral cancers.
Keywords: HPV-associated cancers; biomarkers; circRNAs; squamous cell carcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

