Structure-Function Analyses of New SARS-CoV-2 Variants B.1.1.7, B.1.351 and B.1.1.28.1: Clinical, Diagnostic, Therapeutic and Public Health Implications
- PMID: 33803400
- PMCID: PMC8000172
- DOI: 10.3390/v13030439
Structure-Function Analyses of New SARS-CoV-2 Variants B.1.1.7, B.1.351 and B.1.1.28.1: Clinical, Diagnostic, Therapeutic and Public Health Implications
Abstract
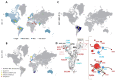
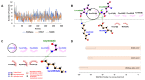
SARS-CoV-2 (Severe Acute Respiratory Syndrome-Coronavirus 2) has accumulated multiple mutations during its global circulation. Recently, three SARS-CoV-2 lineages, B.1.1.7 (501Y.V1), B.1.351 (501Y.V2) and B.1.1.28.1 (P.1), have emerged in the United Kingdom, South Africa and Brazil, respectively. Here, we have presented global viewpoint on implications of emerging SARS-CoV-2 variants based on structural-function impact of crucial mutations occurring in its spike (S), ORF8 and nucleocapsid (N) proteins. While the N501Y mutation was observed in all three lineages, the 501Y.V1 and P.1 accumulated a different set of mutations in the S protein. The missense mutational effects were predicted through a COVID-19 dedicated resource followed by atomistic molecular dynamics simulations. Current findings indicate that some mutations in the S protein might lead to higher affinity with host receptors and resistance against antibodies, but not all are due to different antibody binding (epitope) regions. Mutations may, however, result in diagnostic tests failures and possible interference with binding of newly identified anti-viral candidates against SARS-CoV-2, likely necessitating roll out of recurring "flu-like shots" annually for tackling COVID-19. The functional relevance of these mutations has been described in terms of modulation of host tropism, antibody resistance, diagnostic sensitivity and therapeutic candidates. Besides global economic losses, post-vaccine reinfections with emerging variants can have significant clinical, therapeutic and public health impacts.
Keywords: 501Y.V1; 501Y.V2; B.1.1.28.1; B.1.1.7; B.1.351; COVID-19 vaccines; Clade G; D614G variant; ORF8; P.1; furin cleavage site; immune escape; public health strategies; spike protein; vaccine delivery.
Conflict of interest statement
Authors declare no conflict of interest.
Figures



References
-
- Sironi M., Hasnain S.E., Rosenthal B., Phan T., Luciani F., Shaw M.A., Sallum M.A., Mirhashemi M.E., Morand S., Gonzalez-Candelas F., et al. SARS-CoV-2 and COVID-19: A genetic, epidemiological, and evolutionary perspective. Infect. Genet. Evol. 2020;84:104384. doi: 10.1016/j.meegid.2020.104384. - DOI - PMC - PubMed
-
- Guan Q., Sadykov M., Mfarrej S., Hala S., Naeem R., Nugmanova R., Al-Omari A., Salih S., Mutair A.A., Carr M.J., et al. A genetic barcode of SARS-CoV-2 for monitoring global distribution of different clades during the COVID-19 pandemic. Int. J. Infect. Dis. 2020;100:216–223. doi: 10.1016/j.ijid.2020.08.052. - DOI - PMC - PubMed
-
- Singh H., Singh J., Khubaib M., Jamal S., Sheikh J.A., Kohli S., Hasnain S.E., Rahman S.A. Mapping the genomic landscape & diversity of COVID-19 based on >3950 clinical isolates of SARS-CoV-2: Likely origin & transmission dynamics of isolates sequenced in India. Indian J. Med. Res. 2020;151:474–478. doi: 10.4103/ijmr.IJMR_1253_20. - DOI - PMC - PubMed
-
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

