MYB-Mediated Regulation of Anthocyanin Biosynthesis
- PMID: 33803587
- PMCID: PMC8002911
- DOI: 10.3390/ijms22063103
MYB-Mediated Regulation of Anthocyanin Biosynthesis
Abstract
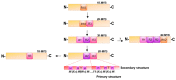
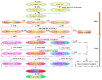
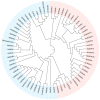
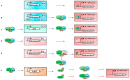
Anthocyanins are natural water-soluble pigments that are important in plants because they endow a variety of colors to vegetative tissues and reproductive plant organs, mainly ranging from red to purple and blue. The colors regulated by anthocyanins give plants different visual effects through different biosynthetic pathways that provide pigmentation for flowers, fruits and seeds to attract pollinators and seed dispersers. The biosynthesis of anthocyanins is genetically determined by structural and regulatory genes. MYB (v-myb avian myeloblastosis viral oncogene homolog) proteins are important transcriptional regulators that play important roles in the regulation of plant secondary metabolism. MYB transcription factors (TFs) occupy a dominant position in the regulatory network of anthocyanin biosynthesis. The TF conserved binding motifs can be combined with other TFs to regulate the enrichment and sedimentation of anthocyanins. In this study, the regulation of anthocyanin biosynthetic mechanisms of MYB-TFs are discussed. The role of the environment in the control of the anthocyanin biosynthesis network is summarized, the complex formation of anthocyanins and the mechanism of environment-induced anthocyanin synthesis are analyzed. Some prospects for MYB-TF to modulate the comprehensive regulation of anthocyanins are put forward, to provide a more relevant basis for further research in this field, and to guide the directed genetic modification of anthocyanins for the improvement of crops for food quality, nutrition and human health.
Keywords: MBW complexes; MYB; anthocyanin; environment; negative regulation; positive regulation; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

