Differential Antimycotic and Antioxidant Potentials of Chemically Synthesized Zinc-Based Nanoparticles Derived from Different Reducing/Complexing Agents against Pathogenic Fungi of Maize Crop
- PMID: 33803825
- PMCID: PMC8003151
- DOI: 10.3390/jof7030223
Differential Antimycotic and Antioxidant Potentials of Chemically Synthesized Zinc-Based Nanoparticles Derived from Different Reducing/Complexing Agents against Pathogenic Fungi of Maize Crop
Abstract
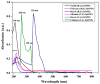
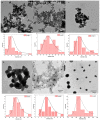
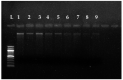
The present study aimed for the synthesis, characterization, and comparative evaluation of anti-oxidant and anti-fungal potentials of zinc-based nanoparticles (ZnNPs) by using different reducing or organic complexing-capping agents. The synthesized ZnNPs exhibited quasi-spherical to hexagonal shapes with average particle sizes ranging from 8 to 210 nm. The UV-Vis spectroscopy of the prepared ZnNPs showed variation in the appearance of characteristic absorption peak(s) for the various reducing/complexing agents i.e., 210 (NaOH and NaBH4), 220 (albumin, and thiourea), 260 and 330 (starch), and 351 nm (cellulose) for wavelengths spanning over 190-800 nm. The FT-IR spectroscopy of the synthesized ZnNPs depicted the functional chemical group diversity. On comparing the antioxidant potential of these ZnNPs, NaOH as reducing agent, (NaOH (RA)) derived ZnNPs presented significantly higher DPPH radical scavenging potential compared to other ZnNPs. The anti-mycotic potential of the ZnNPs as performed through an agar well diffusion assay exhibited variability in the extent of inhibition of the fungal mycelia with maximum inhibition at the highest concentration (40 mg L-1). The NaOH (RA)-derived ZnNPs showcased maximum mycelial inhibition compared to other ZnNPs. Further, incubation of the total genomic DNA with the most effective NaOH (RA)-derived ZnNPs led to intercalation or disintegration of the DNA of all the three fungal pathogens of maize with maximum DNA degrading effect on Macrophomina phaseolina genomic DNA. This study thus identified that differences in size and surface functionalization with the protein (albumin)/polysaccharides (starch, cellulose) diminishes the anti-oxidant and anti-mycotic potential of the generated ZnNPs. However, the NaOH emerged as the best reducing agent for the generation of uniform nano-scale ZnNPs which possessed comparably greater anti-oxidant and antimycotic activities against the three test maize pathogenic fungal cultures.
Keywords: metal oxides; nano-fungicides; pathogenic fungi; protein profiling; radical scavenging activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Chaloner T.M., Gurr S.J., Bebber D.P. The global burden of plant disease tracks crop yields under climate change. bioRxiv. 2020 doi: 10.1101/2020.04.28.066233. - DOI
-
- Elad Y., Pertot I. Climate Change Impacts on Plant Pathogens and Plant Diseases. J. Crop Improv. 2014;28:99–139. doi: 10.1080/15427528.2014.865412. - DOI
-
- Ruffo M.L., Gentry L.F., Henninger A.S., Seebauer J.R., Below F.E. Evaluating management factor contributions to reduce corn yield gaps. Agron. J. 2015;107:495–505. doi: 10.2134/agronj14.0355. - DOI
-
- Mueller D.S., Pathology P., Wise K.A., Bosley D.B., State C., Collins F., Bradley C.A., Pathology P. Corn Yield Loss Estimates Due to Diseases in the United States and Ontario, Canada from 2012 to 2015. Plant Health Prog. 2016;17:211–222. doi: 10.1094/PHP-RS-16-0030. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

