Molecular Assay Development to Monitor the Kinetics of Viable Populations of Two Biocontrol Agents, Bacillus subtilis QST 713 and Gliocladium catenulatum J1446, in the Phyllosphere of Lettuce Leaves
- PMID: 33804029
- PMCID: PMC8001495
- DOI: 10.3390/biology10030224
Molecular Assay Development to Monitor the Kinetics of Viable Populations of Two Biocontrol Agents, Bacillus subtilis QST 713 and Gliocladium catenulatum J1446, in the Phyllosphere of Lettuce Leaves
Abstract
Optimising the use of biocontrol agents (BCAs) requires the temporal tracking of viable populations in the crop phyllosphere to ensure that effective control can be achieved. No sensitive systems for quantifying viable populations of commercially available BCAs, such as Bacillus subtilis and Gliocladium catenulatum, in the phyllosphere of crop plants are available. The objective of this study was to develop a method to quantify viable populations of these two BCAs in the crop phyllosphere. A molecular tool based on propidium monoazide (PMA) (PMAxx™-qPCR) capable of quantifying viable populations of these two BCAs was developed. Samples were treated with PMAxx™ (12.5-100 μM), followed by 15 min incubation, exposure to a 800 W halogen light for 30 min, DNA extraction, and quantification using qPCR. This provided a platform for using the PMAxx™-qPCR technique for both BCAs to differentiate viable from dead cells. The maximum number of dead cells blocked, based on the DNA, was 3.44 log10 for B. subtilis and 5.75 log10 for G. catenulatum. Validation studies showed that this allowed accurate quantification of viable cells. This method provided effective quantification of the temporal changes in viable populations of the BCAs in commercial formulations on lettuce leaves in polytunnel and glasshouse production systems.
Keywords: PMA; biocontrol; commercial formulations; phyllosphere; qPCR; quantitative method; viable cells.
Conflict of interest statement
There are no conflicts of interest.
Figures
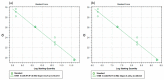
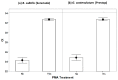

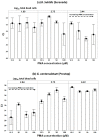



References
-
- Turano A., Pirali F. Laboratory Diagnosis of Infectious Diseases. Springer; Berlin/Heidelberg, Germany: 1988. Quantification Methods in Microbiology.
-
- Sanzani S.M., Li Destri Nicosia M.G., Faedda R., Cacciola S.O., Schena L. Use of quantitative PCR detection methods to study biocontrol agents and phytopathogenic fungi and oomycetes in environmental samples. J. Phytopath. 2014;162:1–13. doi: 10.1111/jph.12147. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

