HERC1 Regulates Breast Cancer Cells Migration and Invasion
- PMID: 33804079
- PMCID: PMC8061768
- DOI: 10.3390/cancers13061309
HERC1 Regulates Breast Cancer Cells Migration and Invasion
Abstract
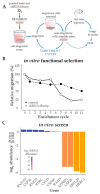
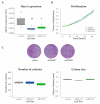
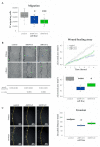
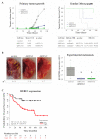
Tumor cell migration and invasion into adjacent tissues is one of the hallmarks of cancer and the first step towards secondary tumors formation, which represents the leading cause of cancer-related deaths. This process is considered an unmet clinical need in the treatment of this disease, particularly in breast cancers characterized by high aggressiveness and metastatic potential. To identify and characterize genes with novel functions as regulators of tumor cell migration and invasion, we performed a genetic loss-of-function screen using a shRNA library directed against the Ubiquitin Proteasome System (UPS) in a highly invasive breast cancer derived cell line. Among the candidates, we validated HERC1 as a gene regulating cell migration and invasion. Furthermore, using animal models, our results indicate that HERC1 silencing affects primary tumor growth and lung colonization. Finally, we conducted an in silico analysis using publicly available protein expression data and observed an inverse correlation between HERC1 expression levels and breast cancer patients' overall survival. Altogether, our findings demonstrate that HERC1 might represent a novel therapeutic target for the development or improvement of breast cancer treatment.
Keywords: HERC1; breast; cancer; invasion; target.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

