Isolation and Characterisation of Bacteriophages with Activity against Invasive Non-Typhoidal Salmonella Causing Bloodstream Infection in Malawi
- PMID: 33804216
- PMCID: PMC7999457
- DOI: 10.3390/v13030478
Isolation and Characterisation of Bacteriophages with Activity against Invasive Non-Typhoidal Salmonella Causing Bloodstream Infection in Malawi
Abstract
In recent years, novel lineages of invasive non-typhoidal Salmonella (iNTS) serovars Typhimurium and Enteritidis have been identified in patients with bloodstream infection in Sub-Saharan Africa. Here, we isolated and characterised 32 phages capable of infecting S. Typhimurium and S. Enteritidis, from water sources in Malawi and the UK. The phages were classified in three major phylogenetic clusters that were geographically distributed. In terms of host range, Cluster 1 phages were able to infect all bacterial hosts tested, whereas Clusters 2 and 3 had a more restricted profile. Cluster 3 contained two sub-clusters, and 3.b contained the most novel isolates. This study represents the first exploration of the potential for phages to target the lineages of Salmonella that are responsible for bloodstream infections in Sub-Saharan Africa.
Keywords: Enteritidis; Malawi; Typhimurium; environmental phage.
Conflict of interest statement
Ella V. Rodwell is affiliated to the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at University of Liverpool in partnership with Public Health England (PHE), in collaboration with University of Warwick. Ella V. Rodwell is based at Public Health England. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health and Social Care or Public Health England. The other authors declare no conflict of interest.
Figures
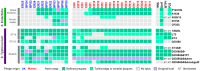



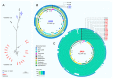
References
-
- Stanaway J.D., Parisi A., Sarkar K., Blacker B.F., Reiner R.C., Hay S.I., Nixon M.R., Dolecek C., James S.L., Mokdad A.H., et al. The global burden of non-typhoidal Salmonella invasive disease: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019;19:1312–1324. doi: 10.1016/S1473-3099(19)30418-9. - DOI - PMC - PubMed
-
- Okoro C.K., Kingsley R.A., Quail M.A., Kankwatira A.M., Feasey N.A., Parkhill J., Dougan G., Gordon M.A. High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal salmonella typhimurium disease. Clin. Infect. Dis. 2012;54:955–963. doi: 10.1093/cid/cir1032. - DOI - PMC - PubMed
-
- Feasey N.A., Hadfield J., Keddy K.H., Dallman T.J., Jacobs J., Deng X., Wigley P., Barquist Barquist L., Langridge G.C., Feltwell T., et al. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat. Genet. 2016;48:1211–1217. doi: 10.1038/ng.3644. - DOI - PMC - PubMed
-
- Kingsley R.A., Msefula C.L., Thomson N.R., Kariuki S., Holt K.E., Gordon M.A., Harris D., Clarke L., Whitehead S., Sangal V., et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. - DOI - PMC - PubMed
-
- Okoro C.K., Kingsley R.A., Connor T.R., Harris S.R., Parry C.M., Al-Mashhadani M.N., Kariuki S., Msefula C.L., Gordon M.A., de Pinna E., et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat. Genet. 2012;44:1215–1221. doi: 10.1038/ng.2423. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

