Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation
- PMID: 33804404
- PMCID: PMC7957543
- DOI: 10.3390/ijms22052474
Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation
Abstract
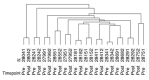
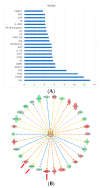
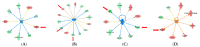
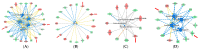
Left Ventricular Outflow Tract (LVOT) obstruction occurs in approximately 70% of Hypertrophic Cardiomyopathy (HCM) patients and currently requires imaging or invasive testing for diagnosis, sometimes in conjunction with provocative physiological or pharmaceutical stimuli. To identify potential biomarkers of LVOT obstruction, we performed proteomics profiling of 1305 plasma proteins in 12 HCM patients with documented LVOT obstruction, referred for surgical myectomy. Plasma was collected at the surgical preoperative visit, approximately one month prior to surgery and then at the post-surgical visit, approximately 3 months later. Proteomic profiles were generated using the aptamer-based SOMAscan assay. Principal Component Analysis using the highest statistically significant proteins separated all preoperative samples from all postoperative samples. Further analysis revealed a set of 25 proteins that distinguished the preoperative and postoperative states with a paired t-test p-value of <0.01. Ingenuity Pathway analysis facilitated the generation of protein interaction networks and the elucidation of key upstream regulators of differentially expressed proteins, such as interferon-γ, TGF-β1, and TNF. Biological pathways affected by surgery included organ inflammation, migration, and motility of leukocytes, fibrosis, vasculogenesis, angiogenesis, acute coronary events, endothelial proliferation, eicosanoid metabolism, calcium flux, apoptosis, and morphology of the cardiovascular system. Our results indicate that surgical relief of dynamic outflow tract obstruction in HCM patients is associated with unique alterations in plasma proteomic profiles that likely reflect improvement in organ inflammation and physiological function.
Keywords: aptamer; cardiovascular disease; hypertrophic cardiomyopathy; myectomy surgery; proteomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Hathout Y., Brody E., Clemens P.R., Cripe L., DeLisle R.K., Furlong P., Gordish-Dressman H., Hache L., Henricson E., Hoffman E.P., et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2015;112:7153–7158. doi: 10.1073/pnas.1507719112. - DOI - PMC - PubMed
-
- Ngo D., Sinha S., Shen D., Kuhn E.W., Keyes M.J., Shi X., Benson M.D., O’Sullivan J.F., Keshishian H., Farrell L.A., et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation. 2016;134:270–285. doi: 10.1161/CIRCULATIONAHA.116.021803. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

