CENPE Inhibition Leads to Mitotic Catastrophe and DNA Damage in Medulloblastoma Cells
- PMID: 33804489
- PMCID: PMC7957796
- DOI: 10.3390/cancers13051028
CENPE Inhibition Leads to Mitotic Catastrophe and DNA Damage in Medulloblastoma Cells
Abstract
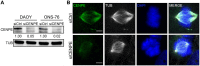
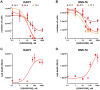
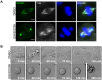
Medulloblastoma (MB) is the most frequent brain tumor in children. The standard treatment consists in surgery, followed by radiotherapy and chemotherapy. These therapies are only partially effective since many patients still die and those who survive suffer from neurological and endocrine disorders. Therefore, more effective therapies are needed. Primary microcephaly (MCPH) is a rare disorder caused by mutations in 25 different genes. Centromere-associated protein E (CENPE) heterozygous mutations cause the MCPH13 syndrome. As for other MCPH genes, CENPE is required for normal proliferation and survival of neural progenitors. Since there is evidence that MB shares many molecular features with neural progenitors, we hypothesized that CENPE could be an effective target for MB treatment. In ONS-76 and DAOY cells, CENPE knockdown induced mitotic defects and apoptosis. Moreover, CENPE depletion induced endogenous DNA damage accumulation, activating TP53 or TP73 as well as cell death signaling pathways. To consolidate CENPE as a target for MB treatment, we tested GSK923295, an allosteric inhibitor already in clinical trial for other cancer types. GSK923295, induced effects similar to CENPE depletion with higher penetrance, at low nM levels, suggesting that CENPE's inhibition could be a therapeutic strategy for MB treatment.
Keywords: 53BP1; CENPE; DNA damage; childhood brain tumor; microcephaly; mitotic catastrophe; γH2AX.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Triscott J., Lee C., Foster C., Manoranjan B., Pambid M.R., Berns R., Fotovati A., Venugopal C., O’Halloran K., Narendran A., et al. Personalizing the Treatment of Pediatric Medulloblastoma: Polo-like Kinase 1 as a Molecular Target in High-Risk Children. Cancer Res. 2013;73:6734–6744. doi: 10.1158/0008-5472.CAN-12-4331. - DOI - PubMed
-
- Cho Y.-J., Tsherniak A., Tamayo P., Santagata S., Ligon A., Greulich H., Berhoukim R., Amani V., Goumnerova L., Eberhart C.G., et al. Integrative Genomic Analysis of Medulloblastoma Identifies a Molecular Subgroup That Drives Poor Clinical Outcome. J. Clin. Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. - DOI - PMC - PubMed
-
- Kool M., Korshunov A., Remke M., Jones D.T.W., Schlanstein M., Northcott P.A., Cho Y.-J., Koster J., Schouten-van Meeteren A., van Vuurden D., et al. Molecular Subgroups of Medulloblastoma: An International Meta-Analysis of Transcriptome, Genetic Aberrations, and Clinical Data of WNT, SHH, Group 3, and Group 4 Medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

