Insights into the Role of Transcriptional Gene Silencing in Response to Herbicide-Treatments in Arabidopsis thaliana
- PMID: 33804990
- PMCID: PMC8037345
- DOI: 10.3390/ijms22073314
Insights into the Role of Transcriptional Gene Silencing in Response to Herbicide-Treatments in Arabidopsis thaliana
Abstract
Herbicide resistance is broadly recognized as the adaptive evolution of weed populations to the intense selection pressure imposed by the herbicide applications. Here, we tested whether transcriptional gene silencing (TGS) and RNA-directed DNA Methylation (RdDM) pathways modulate resistance to commonly applied herbicides. Using Arabidopsis thaliana wild-type plants exposed to sublethal doses of glyphosate, imazethapyr, and 2,4-D, we found a partial loss of TGS and increased susceptibility to herbicides in six out of 11 tested TGS/RdDM mutants. Mutation in REPRESSOR OF SILENCING 1 (ROS1), that plays an important role in DNA demethylation, leading to strongly increased susceptibility to all applied herbicides, and imazethapyr in particular. Transcriptomic analysis of the imazethapyr-treated wild type and ros1 plants revealed a relation of the herbicide upregulated genes to chemical stimulus, secondary metabolism, stress condition, flavonoid biosynthesis, and epigenetic processes. Hypersensitivity to imazethapyr of the flavonoid biosynthesis component TRANSPARENT TESTA 4 (TT4) mutant plants strongly suggests that ROS1-dependent accumulation of flavonoids is an important mechanism for herbicide stress response in A. thaliana. In summary, our study shows that herbicide treatment affects transcriptional gene silencing pathways and that misregulation of these pathways makes Arabidopsis plants more sensitive to herbicide treatment.
Keywords: 2,4-D; ROS1; chromatin mutants; epigenetics; glyphosate; herbicide resistance; imazethapyr.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
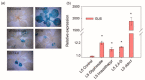



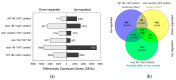

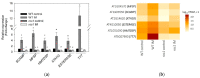

References
-
- Oerke E.C. Crop losses to pests. J. Agric. Sci. 2006;144:31–43. doi: 10.1017/S0021859605005708. - DOI
-
- Heap I. The International Survey of Herbicide Resistant Weeds. [(accessed on 17 January 2021)]; Available online: www.weedscience.org.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

