Pseudomonas Flagella: Generalities and Specificities
- PMID: 33805191
- PMCID: PMC8036289
- DOI: 10.3390/ijms22073337
Pseudomonas Flagella: Generalities and Specificities
Abstract
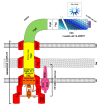
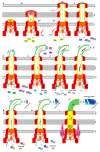
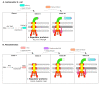
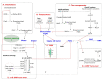
Flagella-driven motility is an important trait for bacterial colonization and virulence. Flagella rotate and propel bacteria in liquid or semi-liquid media to ensure such bacterial fitness. Bacterial flagella are composed of three parts: a membrane complex, a flexible-hook, and a flagellin filament. The most widely studied models in terms of the flagellar apparatus are E. coli and Salmonella. However, there are many differences between these enteric bacteria and the bacteria of the Pseudomonas genus. Enteric bacteria possess peritrichous flagella, in contrast to Pseudomonads, which possess polar flagella. In addition, flagellar gene expression in Pseudomonas is under a four-tiered regulatory circuit, whereas enteric bacteria express flagellar genes in a three-step manner. Here, we use knowledge of E. coli and Salmonella flagella to describe the general properties of flagella and then focus on the specificities of Pseudomonas flagella. After a description of flagellar structure, which is highly conserved among Gram-negative bacteria, we focus on the steps of flagellar assembly that differ between enteric and polar-flagellated bacteria. In addition, we summarize generalities concerning the fuel used for the production and rotation of the flagellar macromolecular complex. The last part summarizes known regulatory pathways and potential links with the type-six secretion system (T6SS).
Keywords: Pseudomonas; T6SS; flagella; flagellar crosstalk.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the writing of the manuscript.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

