High Diversity of β-Glucosidase-Producing Bacteria and Their Genes Associated with Scleractinian Corals
- PMID: 33805379
- PMCID: PMC8037212
- DOI: 10.3390/ijms22073523
High Diversity of β-Glucosidase-Producing Bacteria and Their Genes Associated with Scleractinian Corals
Abstract
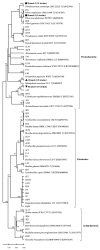
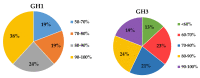
β-Glucosidase is a microbial cellulose multienzyme that plays an important role in the regulation of the entire cellulose hydrolysis process, which is the rate-limiting step in bacterial carbon cycling in marine environments. Despite its importance in coral reefs, the diversity of β-glucosidase-producing bacteria, their genes, and enzymatic characteristics are poorly understood. In this study, 87 β-glucosidase-producing cultivable bacteria were screened from 6 genera of corals. The isolates were assigned to 21 genera, distributed among three groups: Proteobacteria, Firmicutes, and Actinobacteria. In addition, metagenomics was used to explore the genetic diversity of bacterial β-glucosidase enzymes associated with scleractinian corals, which revealed that these enzymes mainly belong to the glycosidase hydrolase family 3 (GH3). Finally, a novel recombinant β-glucosidase, referred to as Mg9373, encompassing 670 amino acids and a molecular mass of 75.2 kDa, was classified as a member of the GH3 family and successfully expressed and characterized. Mg9373 exhibited excellent tolerance to ethanol, NaCl, and glucose. Collectively, these results suggest that the diversity of β-glucosidase-producing bacteria and genes associated with scleractinian corals is high and novel, indicating great potential for applications in the food industry and agriculture.
Keywords: cultivable bacteria; diversity; metagenomic approach; scleractinian corals; β-glucosidase.
Conflict of interest statement
All authors edited and approved the manuscript. The authors declare no conflict of interest.
Figures






References
-
- Forest R., Victor S., Farooq A., Nancy K. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002;243:1–10.
-
- Eugene R. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007;5:355–362. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

