Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice
- PMID: 33805566
- PMCID: PMC8067039
- DOI: 10.3390/life11040291
Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice
Abstract
Rice plants often encounter high temperature stress, but the associated coping strategies are poorly understood. It is known that a prior shorter exposure to high temperature, called thermo-priming, generally results in better adaptation of the plants to subsequent exposure to high temperature stress. High throughput sequencing of transcript and small RNA libraries of rice seedlings primed with short exposure to high temperature followed by high temperature stress and from plants exposed to high temperature without priming was performed. This identified a number of transcripts and microRNAs (miRs) that are induced or down regulated. Among them osa-miR531b, osa-miR5149, osa-miR168a-5p, osa-miR1846d-5p, osa-miR5077, osa-miR156b-3p, osa-miR167e-3p and their respective targets, coding for heat shock activators and repressors, showed differential expression between primed and non-primed plants. These findings were further validated by qRT-PCR. The results indicate that the miR-regulated heat shock proteins (HSPs)/heat shock transcription factors (HSFs) may serve as important regulatory nodes which are induced during thermo-priming for plant survival and development under high temperatures.
Keywords: HSF; HSP; RNA-seq; high temperature stress; interactome; microRNA; thermo-priming.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


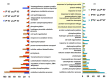

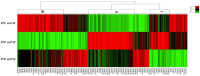


References
-
- Hasanuzzaman M., Nahar K., Fujita M. Extreme temperature responses, oxidative stress and antioxidant defense in plants. In: Vahdati K., Leslie C., editors. Abiotic Stress Plant Responses Appllications in Agriculture. IntechOpen; London, UK: 2013. pp. 169–205.
-
- Teixeira E.I., Fischer G., Van Velthuizen H., Walter C., Ewert F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013;170:206–215. doi: 10.1016/j.agrformet.2011.09.002. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

