Molecular Regulation of Lipogenesis, Adipogenesis and Fat Deposition in Chicken
- PMID: 33805667
- PMCID: PMC8002044
- DOI: 10.3390/genes12030414
Molecular Regulation of Lipogenesis, Adipogenesis and Fat Deposition in Chicken
Abstract
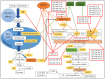
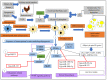
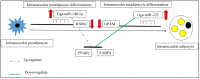
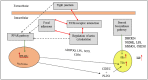
In the poultry industry, excessive fat deposition is considered an undesirable factor, affecting feed efficiency, meat production cost, meat quality, and consumer's health. Efforts to reduce fat deposition in economically important animals, such as chicken, can be made through different strategies; including genetic selection, feeding strategies, housing, and environmental strategies, as well as hormone supplementation. Recent investigations at the molecular level have revealed the significant role of the transcriptional and post-transcriptional regulatory networks and their interaction on modulating fat metabolism in chickens. At the transcriptional level, different transcription factors are known to regulate the expression of lipogenic and adipogenic genes through various signaling pathways, affecting chicken fat metabolism. Alternatively, at the post-transcriptional level, the regulatory mechanism of microRNAs (miRNAs) on lipid metabolism and deposition has added a promising dimension to understand the structural and functional regulatory mechanism of lipid metabolism in chicken. Therefore, this review focuses on the progress made in unraveling the molecular function of genes, transcription factors, and more notably significant miRNAs responsible for regulating adipogenesis, lipogenesis, and fat deposition in chicken. Moreover, a better understanding of the molecular regulation of lipid metabolism will give researchers novel insights to use functional molecular markers, such as miRNAs, for selection against excessive fat deposition to improve chicken production efficiency and meat quality.
Keywords: adipogenesis; chicken; fat deposition; lipogenesis; meat quality; post-transcriptional regulation; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Resnyk C.W., Carré W., Wang X., Porter T.E., Simon J., Le Bihan-Duval E., Duclos M.J., Aggrey S.E., Cogburn L.A. Transcriptional Analysis of Abdominal Fat in Genetically Fat and Lean Chickens Reveals Adipokines, Lipogenic Genes and a Link between Hemostasis and Leanness. BMC Genom. 2013;14:1–26. doi: 10.1186/1471-2164-14-557. - DOI - PMC - PubMed
-
- Mm G., Li M. Expression of Adipocyte Fatty Acid-Binding Protein Gene in Abdominal Adipose Tissue and Its Association with Growth and Fatness Traits in Commercial Meat Type Chickens. IMedPub J. 2018;1:1–10.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

