The Pseudo-Circular Genomes of Flaviviruses: Structures, Mechanisms, and Functions of Circularization
- PMID: 33805761
- PMCID: PMC7999817
- DOI: 10.3390/cells10030642
The Pseudo-Circular Genomes of Flaviviruses: Structures, Mechanisms, and Functions of Circularization
Abstract
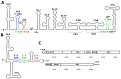
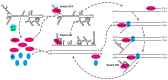
The circularization of viral genomes fulfills various functions, from evading host defense mechanisms to promoting specific replication and translation patterns supporting viral proliferation. Here, we describe the genomic structures and associated host factors important for flaviviruses genome circularization and summarize their functional roles. Flaviviruses are relatively small, single-stranded, positive-sense RNA viruses with genomes of approximately 11 kb in length. These genomes contain motifs at their 5' and 3' ends, as well as in other regions, that are involved in circularization. These motifs are highly conserved throughout the Flavivirus genus and occur both in mature virions and within infected cells. We provide an overview of these sequence motifs and RNA structures involved in circularization, describe their linear and circularized structures, and discuss the proteins that interact with these circular structures and that promote and regulate their formation, aiming to clarify the key features of genome circularization and understand how these affect the flaviviruses life cycle.
Keywords: RNA structure; circular RNA; circular genomes; flavivirus; virus genomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Aedes Albopictus - actsheet for Experts. [(accessed on 6 July 2020)]; Available online: https://www.ecdc.europa.eu/en/disease-vectors/facts/mosquito-factsheets/....
-
- Aedes Aegypti - Factsheet for Experts. [(accessed on 6 July 2020)]; Available online: https://www.ecdc.europa.eu/en/disease-vectors/facts/mosquito-factsheets/....
-
- Dengue and Severe Dengue. [(accessed on 6 July 2020)]; Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
-
- Zika Virus. [(accessed on 6 July 2020)]; Available online: https://www.who.int/news-room/fact-sheets/detail/zika-virus.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

