The Interaction of Human and Epstein-Barr Virus miRNAs with Multiple Sclerosis Risk Loci
- PMID: 33805769
- PMCID: PMC8000127
- DOI: 10.3390/ijms22062927
The Interaction of Human and Epstein-Barr Virus miRNAs with Multiple Sclerosis Risk Loci
Abstract
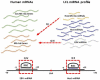
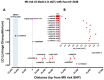
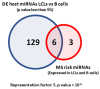
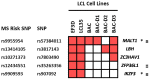
Although the causes of Multiple Sclerosis (MS) still remain largely unknown, multiple lines of evidence suggest that Epstein-Barr virus (EBV) infection may contribute to the development of MS. Here, we aimed to identify the potential contribution of EBV-encoded and host cellular miRNAs to MS pathogenesis. We identified differentially expressed host miRNAs in EBV infected B cells (LCLs) and putative host/EBV miRNA interactions with MS risk loci. We estimated the genotype effect of MS risk loci on the identified putative miRNA:mRNA interactions in silico. We found that the protective allele of MS risk SNP rs4808760 reduces the expression of hsa-mir-3188-3p. In addition, our analysis suggests that hsa-let-7b-5p may interact with ZC3HAV1 differently in LCLs compared to B cells. In vitro assays indicated that the protective allele of MS risk SNP rs10271373 increases ZC3HAV1 expression in LCLs, but not in B cells. The higher expression for the protective allele in LCLs is consistent with increased IFN response via ZC3HAV1 and so decreased immune evasion by EBV. Taken together, this provides evidence that EBV infection dysregulates the B cell miRNA machinery, including MS risk miRNAs, which may contribute to MS pathogenesis via interaction with MS risk genes either directly or indirectly.
Keywords: EBNA2; EBV; GWAS; MS; ZC3HAV1; miRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

