Corynebacterium diphtheriae Proteome Adaptation to Cell Culture Medium and Serum
- PMID: 33805816
- PMCID: PMC8005964
- DOI: 10.3390/proteomes9010014
Corynebacterium diphtheriae Proteome Adaptation to Cell Culture Medium and Serum
Abstract
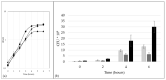
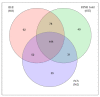
Host-pathogen interactions are often studied in vitro using primary or immortal cell lines. This set-up avoids ethical problems of animal testing and has the additional advantage of lower costs. However, the influence of cell culture media on bacterial growth and metabolism is not considered or investigated in most cases. To address this question growth and proteome adaptation of Corynebacterium diphtheriae strain ISS3319 were investigated in this study. Bacteria were cultured in standard growth medium, cell culture medium, and fetal calf serum. Mass spectrometric analyses and label-free protein quantification hint at an increased bacterial pathogenicity when grown in cell culture medium as well as an influence of the growth medium on the cell envelope.
Keywords: diphtheria; host-pathogen interaction; label-free quantification; metabolic pathway; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Burkovski A. Diphtheria and its etiological agents. In: Burkovski A., editor. Corynebacterium Diphtheriae and Related Toxigenic Species. Springer International Publishing; Dordrecht, The Netherlands: 2014. pp. 1–14.
-
- World Health Organization Diphtheria Reported Cases. [(accessed on 16 March 2020)]; Available online: https://apps.who.int/immunization_monitoring/globalsummary/timeseries/ts....
LinkOut - more resources
Full Text Sources
Other Literature Sources

