Systems Approaches to Treatment Response to Imatinib in Severe Asthma: A Pilot Study
- PMID: 33805900
- PMCID: PMC8064376
- DOI: 10.3390/jpm11040240
Systems Approaches to Treatment Response to Imatinib in Severe Asthma: A Pilot Study
Abstract
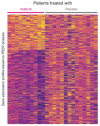
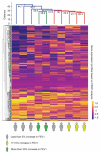
There is an acute need for advances in pharmacologic therapies and a better understanding of novel drug targets for severe asthma. Imatinib, a tyrosine kinase inhibitor, has been shown to improve forced expiratory volume in 1 s (FEV1) in a clinical trial of patients with severe asthma. In a pilot study, we applied systems biology approaches to epithelium gene expression from these clinical trial patients treated with imatinib to better understand lung function response with imatinib treatment. Bronchial brushings from ten imatinib-treated patient samples and 14 placebo-treated patient samples were analyzed. We used personalized perturbation profiles (PEEPs) to characterize gene expression patterns at the individual patient level. We found that strong responders-patients with greater than 20% increase in FEV1-uniquely shared multiple downregulated mitochondrial-related pathways. In comparison, weak responders (5-10% FEV1 increase), and non-responders to imatinib shared none of these pathways. The use of PEEP highlights its potential for application as a systems biology tool to develop individual-level approaches to predicting disease phenotypes and response to treatment in populations needing innovative therapies. These results support a role for mitochondrial pathways in airflow limitation in severe asthma and as potential therapeutic targets in larger clinical trials.
Keywords: asthma subtypes; mitochondria; personalized medicine; personalized perturbation profiles; pharmacogenetics; systems biology.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. S.H.B., D.F., E.M., A.R. have no relevant conflict of interest to disclose. K.N.C. has served as an advisory board member for TEVA, Regeneron, Novartis, Optinose, GlaxoSmithKline, Sanofi-Pasteur, Genentech, and Blueprint Medicines and reports personal fees from Novartis, Third Harmonic, and Ribon Therapeutics. E.I. reports personal fees from AB Science, Allergy and Asthma Network (AAN PCORI), Biometry, Equillium, Merck, NHLBI, 4D Pharma, Pneuma Respiratory, PPS Health, Regeneron, Sanofi Genzyme, Sienna Biopharmaceutical, grants, personal fees and non-financial support from Genentech, grants and personal fees from Amgen, AstraZeneca, Avillion, Novartis, personal fees and non-financial support from GlaxoSmithKline, TEVA, grants from Gossamer Bio, grants and non-financial support from Circassia, non-financial support from Boehringer Ingelheim, other from Vorso Corp, outside the submitted work. J.A.B. has served as a scientific advisory board member for Sanofi-Regeneron, Third Harmonic Bio, and Siolata Therapeutics. S.T.W. reports receiving authorship payment from UpToDate. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Centers for Disease Control and Prevention AsthmaStats: Uncontrolled Asthma Among Adults. [(accessed on 7 April 2020)];2019 Available online: https://www.cdc.gov/asthma/asthma_stats/default.htm.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

