Mutation in the Ciliary Protein C2CD3 Reveals Organ-Specific Mechanisms of Hedgehog Signal Transduction in Avian Embryos
- PMID: 33805906
- PMCID: PMC8103285
- DOI: 10.3390/jdb9020012
Mutation in the Ciliary Protein C2CD3 Reveals Organ-Specific Mechanisms of Hedgehog Signal Transduction in Avian Embryos
Abstract
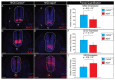
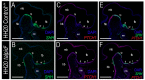
Primary cilia are ubiquitous microtubule-based organelles that serve as signaling hubs for numerous developmental pathways, most notably the Hedgehog (Hh) pathway. Defects in the structure or function of primary cilia result in a class of diseases called ciliopathies. It is well known that primary cilia participate in transducing a Hh signal, and as such ciliopathies frequently present with phenotypes indicative of aberrant Hh function. Interestingly, the exact mechanisms of cilia-dependent Hh signaling transduction are unclear as some ciliopathic animal models simultaneously present with gain-of-Hh phenotypes in one organ system and loss-of-Hh phenotypes in another. To better understand how Hh signaling is perturbed across different tissues in ciliopathic conditions, we examined four distinct Hh-dependent signaling centers in the naturally occurring avian ciliopathic mutant talpid2 (ta2). In addition to the well-known and previously reported limb and craniofacial malformations, we observed dorsal-ventral patterning defects in the neural tube, and a shortened gastrointestinal tract. Molecular analyses for elements of the Hh pathway revealed that the loss of cilia impact transduction of an Hh signal in a tissue-specific manner at variable levels of the pathway. These studies will provide increased knowledge into how impaired ciliogenesis differentially regulates Hh signaling across tissues and will provide potential avenues for future targeted therapeutic treatments.
Keywords: C2CD3; Hedgehog signaling; ciliopathies; craniofacial; hindgut; limb; neural tube; primary cilia; talpid2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

