A S imple, A ffordable, R apid, S tabilized, Co lorimetric, V ersatile RT-LAMP Assay to Detect SARS-CoV-2
- PMID: 33806456
- PMCID: PMC8000859
- DOI: 10.3390/diagnostics11030438
A S imple, A ffordable, R apid, S tabilized, Co lorimetric, V ersatile RT-LAMP Assay to Detect SARS-CoV-2
Abstract
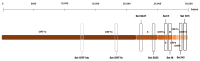
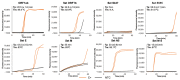
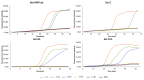
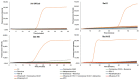
The SARS-CoV-2 pandemic has forced all countries worldwide to rapidly develop and implement widespread testing to control and manage the Coronavirus Disease 2019 (COVID-19). reverse-transcription (RT)-qPCR is the gold standard molecular diagnostic method for COVID-19, mostly in automated testing platforms. These systems are accurate and effective, but also costly, time-consuming, high-technological, infrastructure-dependent, and currently suffer from commercial reagent supply shortages. The reverse-transcription loop-mediated isothermal amplification (RT-LAMP) can be used as an alternative testing method. Here, we present a novel versatile (real-time and colorimetric) RT-LAMP for the simple (one-step), affordable (~1.7 €/sample), and rapid detection of SARS-CoV-2 targeting both ORF1ab and N genes of the novel virus genome. We demonstrate the assay on RT-qPCR-positive clinical samples, obtaining most positive results under 25 min. In addition, a novel 30-min one-step drying protocol has been developed to stabilize the RT-LAMP reaction mixtures, allowing them to be stored at room temperature functionally for up to two months, as predicted by the Q10. This Dry-RT-LAMP methodology is suitable for potentially ready-to-use COVID-19 diagnosis. After further testing and validation, it could be easily applied both in developed and in low-income countries yielding rapid and reliable results.
Keywords: RT-LAMP; SARS-CoV-2; dry-RT-LAMP; molecular diagnostics; point-of-care.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

