Stoichiometric Thiol Redox Proteomics for Quantifying Cellular Responses to Perturbations
- PMID: 33807006
- PMCID: PMC8004825
- DOI: 10.3390/antiox10030499
Stoichiometric Thiol Redox Proteomics for Quantifying Cellular Responses to Perturbations
Abstract
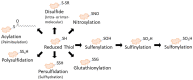
Post-translational modifications regulate the structure and function of proteins that can result in changes to the activity of different pathways. These include modifications altering the redox state of thiol groups on protein cysteine residues, which are sensitive to oxidative environments. While mass spectrometry has advanced the identification of protein thiol modifications and expanded our knowledge of redox-sensitive pathways, the quantitative aspect of this technique is critical for the field of redox proteomics. In this review, we describe how mass spectrometry-based redox proteomics has enabled researchers to accurately quantify the stoichiometry of reversible oxidative modifications on specific cysteine residues of proteins. We will describe advancements in the methodology that allow for the absolute quantitation of thiol modifications, as well as recent reports that have implemented this approach. We will also highlight the significance and application of such measurements and why they are informative for the field of redox biology.
Keywords: cysteine; redox post-translational modifications; redox proteomics; thiol proteome.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

