Molecular Signatures of Natural Killer Cells in CMV-Associated Anterior Uveitis, A New Type of CMV-Induced Disease in Immunocompetent Individuals
- PMID: 33807229
- PMCID: PMC8037729
- DOI: 10.3390/ijms22073623
Molecular Signatures of Natural Killer Cells in CMV-Associated Anterior Uveitis, A New Type of CMV-Induced Disease in Immunocompetent Individuals
Abstract
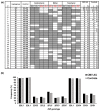
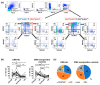
Cytomegalovirus (CMV) causes clinical issues primarily in immune-suppressed conditions. CMV-associated anterior uveitis (CMV-AU) is a notable new disease entity manifesting recurrent ocular inflammation in immunocompetent individuals. As patient demographics indicated contributions from genetic background and immunosenescence as possible underlying pathological mechanisms, we analyzed the immunogenetics of the cohort in conjunction with cell phenotypes to identify molecular signatures of CMV-AU. Among the immune cell types, natural killer (NK) cells are main responders against CMV. Therefore, we first characterized variants of polymorphic genes that encode differences in CMV-related human NK cell responses (Killer cell Immunoglobulin-like Receptors (KIR) and HLA class I) in 122 CMV-AU patients. The cases were then stratified according to their genetic features and NK cells were analyzed for human CMV-related markers (CD57, KLRG1, NKG2C) by flow cytometry. KIR3DL1 and HLA class I combinations encoding strong receptor-ligand interactions were present at substantially higher frequencies in CMV-AU. In these cases, NK cell profiling revealed expansion of the subset co-expressing CD57 and KLRG1, and together with KIR3DL1 and the CMV-recognizing NKG2C receptor. The findings imply that a mechanism of CMV-AU pathogenesis likely involves CMV-responding NK cells co-expressing CD57/KLRG1/NKG2C that develop on a genetic background of KIR3DL1/HLA-B allotypes encoding strong receptor-ligand interactions.
Keywords: CD57; HLA class I; KLRG1; NKG2C; cytomegalovirus; cytomegalovirus-associated anterior uveitis; killer cell immunoglobulin-like receptors; natural killer cells.
Conflict of interest statement
The authors declare no conflict of interest in this study. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Expansion of a unique CD57⁺NKG2Chi natural killer cell subset during acute human cytomegalovirus infection.Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14725-32. doi: 10.1073/pnas.1110900108. Epub 2011 Aug 8. Proc Natl Acad Sci U S A. 2011. PMID: 21825173 Free PMC article.
-
Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function.Blood. 2012 Mar 15;119(11):2665-74. doi: 10.1182/blood-2011-10-386995. Epub 2011 Dec 16. Blood. 2012. PMID: 22180440 Free PMC article.
-
Regulation of Adaptive NK Cells and CD8 T Cells by HLA-C Correlates with Allogeneic Hematopoietic Cell Transplantation and with Cytomegalovirus Reactivation.J Immunol. 2015 Nov 1;195(9):4524-36. doi: 10.4049/jimmunol.1401990. Epub 2015 Sep 28. J Immunol. 2015. PMID: 26416275 Free PMC article.
-
Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host-pathogen interaction.Eur J Immunol. 2013 May;43(5):1133-41. doi: 10.1002/eji.201243117. Eur J Immunol. 2013. PMID: 23552990 Review.
-
Cytomegalovirus: an unlikely ally in the fight against blood cancers?Clin Exp Immunol. 2018 Sep;193(3):265-274. doi: 10.1111/cei.13152. Clin Exp Immunol. 2018. PMID: 29737525 Free PMC article. Review.
Cited by
-
Clinical Features of Glaucoma Associated with Cytomegalovirus Corneal Endotheliitis.Clin Ophthalmol. 2022 Aug 19;16:2705-2711. doi: 10.2147/OPTH.S376039. eCollection 2022. Clin Ophthalmol. 2022. PMID: 36017508 Free PMC article.
-
Intraocular human cytomegaloviruses of ocular diseases are distinct from those of viremia and are capable of escaping from innate and adaptive immunity by exploiting HLA-E-mediated peripheral and central tolerance.Front Immunol. 2022 Oct 19;13:1008220. doi: 10.3389/fimmu.2022.1008220. eCollection 2022. Front Immunol. 2022. PMID: 36341392 Free PMC article.
-
Assessing the Response of Human NK Cell Subsets to Infection by Clinically Isolated Virus Strains.Methods Mol Biol. 2022;2463:205-220. doi: 10.1007/978-1-0716-2160-8_15. Methods Mol Biol. 2022. PMID: 35344177
-
Allele imputation for the killer cell immunoglobulin-like receptor KIR3DL1/S1.PLoS Comput Biol. 2022 Feb 22;18(2):e1009059. doi: 10.1371/journal.pcbi.1009059. eCollection 2022 Feb. PLoS Comput Biol. 2022. PMID: 35192601 Free PMC article.
References
-
- Sugar E.A., Jabs D.A., Ahuja A., Thorne J.E., Danis R.P., Meinert C.L., Studies of the Ocular Complications of AIDS Research Group Incidence of cytomegalovirus retinitis in the era of highly active antiretroviral therapy. Am. J. Ophthalmol. 2012;153:1016–1024. doi: 10.1016/j.ajo.2011.11.014. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

