Metagenomic Analysis of the Respiratory Microbiome of a Broiler Flock from Hatching to Processing
- PMID: 33807233
- PMCID: PMC8065701
- DOI: 10.3390/microorganisms9040721
Metagenomic Analysis of the Respiratory Microbiome of a Broiler Flock from Hatching to Processing
Abstract
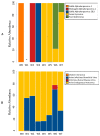
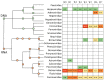
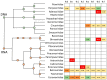
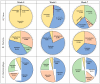
Elucidating the complex microbial interactions in biological environments requires the identification and characterization of not only the bacterial component but also the eukaryotic viruses, bacteriophage, and fungi. In a proof of concept experiment, next generation sequencing approaches, accompanied by the development of novel computational and bioinformatics tools, were utilized to examine the evolution of the microbial ecology of the avian trachea during the growth of a healthy commercial broiler flock. The flock was sampled weekly, beginning at placement and concluding at 49 days, the day before processing. Metagenomic sequencing of DNA and RNA was utilized to examine the bacteria, virus, bacteriophage, and fungal components during flock growth. The utility of using a metagenomic approach to study the avian respiratory virome was confirmed by detecting the dysbiosis in the avian respiratory virome of broiler chickens diagnosed with infection with infectious laryngotracheitis virus. This study provides the first comprehensive analysis of the ecology of the avian respiratory microbiome and demonstrates the feasibility for the use of this approach in future investigations of avian respiratory diseases.
Keywords: avian; bioinformatics; chicken; microbiome; respiratory; virome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript. Or in the decision to publish the results.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

