ARMCX3 Mediates Susceptibility to Hepatic Tumorigenesis Promoted by Dietary Lipotoxicity
- PMID: 33807672
- PMCID: PMC7961652
- DOI: 10.3390/cancers13051110
ARMCX3 Mediates Susceptibility to Hepatic Tumorigenesis Promoted by Dietary Lipotoxicity
Abstract
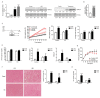
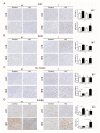
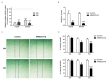
ARMCX3 is encoded by a member of the Armcx gene family and is known to be involved in nervous system development and function. We found that ARMCX3 is markedly upregulated in mouse liver in response to high lipid availability, and that hepatic ARMCX3 is upregulated in patients with NAFLD and hepatocellular carcinoma (HCC). Mice were subjected to ARMCX3 invalidation (inducible ARMCX3 knockout) and then exposed to a high-fat diet and diethylnitrosamine-induced hepatocarcinogenesis. The effects of experimental ARMCX3 knockdown or overexpression in HCC cell lines were also analyzed. ARMCX3 invalidation protected mice against high-fat-diet-induced NAFLD and chemically induced hepatocarcinogenesis. ARMCX3 invalidation promoted apoptotic cell death and macrophage infiltration in livers of diethylnitrosamine-treated mice maintained on a high-fat diet. ARMCX3 downregulation reduced the viability, clonality and migration of HCC cell lines, whereas ARMCX3 overexpression caused the reciprocal effects. SOX9 was found to mediate the effects of ARMCX3 in hepatic cells, with the SOX9 interaction required for the effects of ARMCX3 on hepatic cell proliferation. In conclusion, ARMCX3 is identified as a novel molecular actor in liver physiopathology and carcinogenesis. ARMCX3 downregulation appears to protect against hepatocarcinogenesis, especially under conditions of high dietary lipid-mediated hepatic insult.
Keywords: Alex3; HCC; NAFLD; SOX9; lipotoxicity; obesity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Lopez-Domenech G., Serrat R., Mirra S., D’Aniello S., Somorjai I., Abad A., Vitureira N., García-Arumí E., Alonso M.T., Rodriguez-Prados M., et al. The Eutherian Armcx genes regulate mitochondrial trafficking in neurons and interact with Miro and Trak2. Nat. Commun. 2012;3:814. doi: 10.1038/ncomms1829. - DOI - PubMed
-
- Serrat R., Lopez-Domenech G., Mirra S., Quevedo M., Garcia-Fernandez J., Burgaya F., Soriano E. The non-canonical Wnt/PKC pathway regulates mitochondrial dynamics through degradation of the arm-like domain-containing protein ARMCX3. PLoS ONE. 2013;8:e67773. doi: 10.1371/journal.pone.0067773. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

