CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands
- PMID: 33807682
- PMCID: PMC7961906
- DOI: 10.3390/ijms22052614
CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands
Abstract
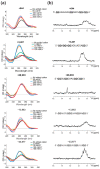
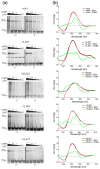
The Coronavirus Disease 2019 (COVID-19) pandemic has become a global health emergency with no effective medical treatment and with incipient vaccines. It is caused by a new positive-sense RNA virus called severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2). G-quadruplexes (G4s) are nucleic acid secondary structures involved in the control of a variety of biological processes including viral replication. Using several G4 prediction tools, we identified highly putative G4 sequences (PQSs) within the positive-sense (+gRNA) and negative-sense (-gRNA) RNA strands of SARS-CoV-2 conserved in related betacoronaviruses. By using multiple biophysical techniques, we confirmed the formation of two G4s in the +gRNA and provide the first evidence of G4 formation by two PQSs in the -gRNA of SARS-CoV-2. Finally, biophysical and molecular approaches were used to demonstrate for the first time that CNBP, the main human cellular protein bound to SARS-CoV-2 RNA genome, binds and promotes the unfolding of G4s formed by both strands of SARS-CoV-2 RNA genome. Our results suggest that G4s found in SARS-CoV-2 RNA genome and its negative-sense replicative intermediates, as well as the cellular proteins that interact with them, are relevant factors for viral genes expression and replication cycle, and may constitute interesting targets for antiviral drugs development.
Keywords: CNBP; COVID-19; G-quadruplex; SARS-CoV-2; coronavirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
-
- Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42:3–11. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

