Drug Discovery in Liver Disease Using Kinome Profiling
- PMID: 33807722
- PMCID: PMC7961955
- DOI: 10.3390/ijms22052623
Drug Discovery in Liver Disease Using Kinome Profiling
Abstract
The liver is one of the most important organs, playing critical roles in maintaining biochemical homeostasis. Accordingly, disease of the liver is often debilitating and responsible for untold human misery. As biochemical nexus, with kinases being master regulators of cellular biochemistry, targeting kinase enzymes is an obvious avenue for treating liver disease. Development of such therapy, however, is hampered by the technical difficulty of obtaining comprehensive insight into hepatic kinase activity, a problem further compounded by the often unique aspects of hepatic kinase activities, which makes extrapolations from other systems difficult. This consideration prompted us to review the current state of the art with respect to kinome profiling approaches towards the hepatic kinome. We observe that currently four different approaches are available, all showing significant promise. Hence we postulate that insight into the hepatic kinome will quickly increase, leading to rational kinase-targeted therapy for different liver diseases.
Keywords: hepatitis; hepatocellular carcinoma; hepatocytes; kinases; kinome; liver; peptide arrays.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
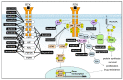
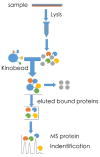
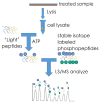
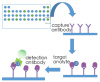
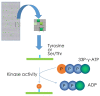
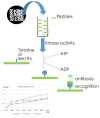
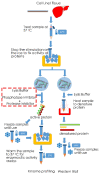
References
-
- Hall J.E. Pocket Companion to Guyton & Hall Textbook of Medical Physiology E-Book. Elsevier Health Sciences; Amsterdam, The Netherlands: 2015.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

