Ensemble of Multiple Classifiers for Multilabel Classification of Plant Protein Subcellular Localization
- PMID: 33808227
- PMCID: PMC8066735
- DOI: 10.3390/life11040293
Ensemble of Multiple Classifiers for Multilabel Classification of Plant Protein Subcellular Localization
Abstract
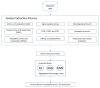
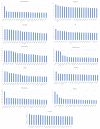
The accurate prediction of protein localization is a critical step in any functional genome annotation process. This paper proposes an improved strategy for protein subcellular localization prediction in plants based on multiple classifiers, to improve prediction results in terms of both accuracy and reliability. The prediction of plant protein subcellular localization is challenging because the underlying problem is not only a multiclass, but also a multilabel problem. Generally, plant proteins can be found in 10-14 locations/compartments. The number of proteins in some compartments (nucleus, cytoplasm, and mitochondria) is generally much greater than that in other compartments (vacuole, peroxisome, Golgi, and cell wall). Therefore, the problem of imbalanced data usually arises. Therefore, we propose an ensemble machine learning method based on average voting among heterogeneous classifiers. We first extracted various types of features suitable for each type of protein localization to form a total of 479 feature spaces. Then, feature selection methods were used to reduce the dimensions of the features into smaller informative feature subsets. This reduced feature subset was then used to train/build three different individual models. In the process of combining the three distinct classifier models, we used an average voting approach to combine the results of these three different classifiers that we constructed to return the final probability prediction. The method could predict subcellular localizations in both single- and multilabel locations, based on the voting probability. Experimental results indicated that the proposed ensemble method could achieve correct classification with an overall accuracy of 84.58% for 11 compartments, on the basis of the testing dataset.
Keywords: average voting; consensus voting; ensemble machine learning; feature extraction; feature selection; go term; plant protein; subcellular localization prediction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Minimalist ensemble algorithms for genome-wide protein localization prediction.BMC Bioinformatics. 2012 Jul 3;13:157. doi: 10.1186/1471-2105-13-157. BMC Bioinformatics. 2012. PMID: 22759391 Free PMC article.
-
Ensemble of heterogeneous classifiers for diagnosis and prediction of coronary artery disease with reduced feature subset.Comput Methods Programs Biomed. 2021 Jan;198:105770. doi: 10.1016/j.cmpb.2020.105770. Epub 2020 Sep 30. Comput Methods Programs Biomed. 2021. PMID: 33027698
-
CE-PLoc: an ensemble classifier for predicting protein subcellular locations by fusing different modes of pseudo amino acid composition.Comput Biol Chem. 2011 Aug 10;35(4):218-29. doi: 10.1016/j.compbiolchem.2011.05.003. Epub 2011 May 27. Comput Biol Chem. 2011. PMID: 21864791
-
Hierarchical ensemble methods for protein function prediction.ISRN Bioinform. 2014 May 4;2014:901419. doi: 10.1155/2014/901419. eCollection 2014. ISRN Bioinform. 2014. PMID: 25937954 Free PMC article. Review.
-
RNA trafficking and subcellular localization-a review of mechanisms, experimental and predictive methodologies.Brief Bioinform. 2023 Sep 20;24(5):bbad249. doi: 10.1093/bib/bbad249. Brief Bioinform. 2023. PMID: 37466130 Free PMC article. Review.
Cited by
-
PlasmidEC and gplas2: an optimized short-read approach to predict and reconstruct antibiotic resistance plasmids in Escherichia coli.Microb Genom. 2024 Feb;10(2):001193. doi: 10.1099/mgen.0.001193. Microb Genom. 2024. PMID: 38376388 Free PMC article.
-
Genome-Wide Identification of Strawberry C2H2-ZFP C1-2i Subclass and the Potential Function of FaZAT10 in Abiotic Stress.Int J Mol Sci. 2022 Oct 28;23(21):13079. doi: 10.3390/ijms232113079. Int J Mol Sci. 2022. PMID: 36361867 Free PMC article.
-
The synthesis of triacylglycerol by diacylglycerol acyltransferases (CsDGAT1A and CsDGAT2D) is essential for tolerance of cucumber's resistance to low-temperature stress.Plant Cell Rep. 2024 Jul 16;43(8):196. doi: 10.1007/s00299-024-03282-z. Plant Cell Rep. 2024. PMID: 39009888
-
Recent Advances in the Prediction of Subcellular Localization of Proteins and Related Topics.Front Bioinform. 2022 May 19;2:910531. doi: 10.3389/fbinf.2022.910531. eCollection 2022. Front Bioinform. 2022. PMID: 36304291 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources