iEnhancer-GAN: A Deep Learning Framework in Combination with Word Embedding and Sequence Generative Adversarial Net to Identify Enhancers and Their Strength
- PMID: 33808317
- PMCID: PMC8036415
- DOI: 10.3390/ijms22073589
iEnhancer-GAN: A Deep Learning Framework in Combination with Word Embedding and Sequence Generative Adversarial Net to Identify Enhancers and Their Strength
Abstract
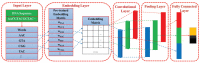
As critical components of DNA, enhancers can efficiently and specifically manipulate the spatial and temporal regulation of gene transcription. Malfunction or dysregulation of enhancers is implicated in a slew of human pathology. Therefore, identifying enhancers and their strength may provide insights into the molecular mechanisms of gene transcription and facilitate the discovery of candidate drug targets. In this paper, a new enhancer and its strength predictor, iEnhancer-GAN, is proposed based on a deep learning framework in combination with the word embedding and sequence generative adversarial net (Seq-GAN). Considering the relatively small training dataset, the Seq-GAN is designed to generate artificial sequences. Given that each functional element in DNA sequences is analogous to a "word" in linguistics, the word segmentation methods are proposed to divide DNA sequences into "words", and the skip-gram model is employed to transform the "words" into digital vectors. In view of the powerful ability to extract high-level abstraction features, a convolutional neural network (CNN) architecture is constructed to perform the identification tasks, and the word vectors of DNA sequences are vertically concatenated to form the embedding matrices as the input of the CNN. Experimental results demonstrate the effectiveness of the Seq-GAN to expand the training dataset, the possibility of applying word segmentation methods to extract "words" from DNA sequences, the feasibility of implementing the skip-gram model to encode DNA sequences, and the powerful prediction ability of the CNN. Compared with other state-of-the-art methods on the training dataset and independent test dataset, the proposed method achieves a significantly improved overall performance. It is anticipated that the proposed method has a certain promotion effect on enhancer related fields.
Keywords: convolutional neural network; enhancer; sequence generative adversarial net; word embedding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Beytebiere J.R., Trott A.J., Greenwell B.J., Osborne C.A., Vitet H., Spence J., Yoo S.-H., Chen Z., Takahashi J.S., Ghaffari N., et al. Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev. 2019;33:294–309. doi: 10.1101/gad.322198.118. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

