GmNF-YC4-2 Increases Protein, Exhibits Broad Disease Resistance and Expedites Maturity in Soybean
- PMID: 33808355
- PMCID: PMC8036377
- DOI: 10.3390/ijms22073586
GmNF-YC4-2 Increases Protein, Exhibits Broad Disease Resistance and Expedites Maturity in Soybean
Abstract
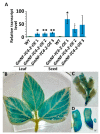
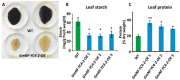
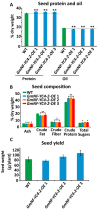
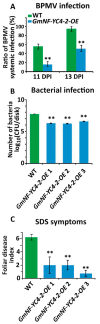
The NF-Y gene family is a highly conserved set of transcription factors. The functional transcription factor complex is made up of a trimer between NF-YA, NF-YB, and NF-YC proteins. While mammals typically have one gene for each subunit, plants often have multigene families for each subunit which contributes to a wide variety of combinations and functions. Soybean plants with an overexpression of a particular NF-YC isoform GmNF-YC4-2 (Glyma.04g196200) in soybean cultivar Williams 82, had a lower amount of starch in its leaves, a higher amount of protein in its seeds, and increased broad disease resistance for bacterial, viral, and fungal infections in the field, similar to the effects of overexpression of its isoform GmNF-YC4-1 (Glyma.06g169600). Interestingly, GmNF-YC4-2-OE (overexpression) plants also filled pods and senesced earlier, a novel trait not found in GmNF-YC4-1-OE plants. No yield difference was observed in GmNF-YC4-2-OE compared with the wild-type control. Sequence alignment of GmNF-YC4-2, GmNF-YC4-1 and AtNF-YC1 indicated that faster maturation may be a result of minor sequence differences in the terminal ends of the protein compared to the closely related isoforms.
Keywords: NF-YC4 transcription factor; disease resistance; early maturation; seed protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

