Untargeted Metabolomics Analysis by UHPLC-MS/MS of Soybean Plant in a Compatible Response to Phakopsora pachyrhizi Infection
- PMID: 33808519
- PMCID: PMC8003322
- DOI: 10.3390/metabo11030179
Untargeted Metabolomics Analysis by UHPLC-MS/MS of Soybean Plant in a Compatible Response to Phakopsora pachyrhizi Infection
Abstract
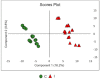
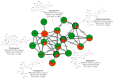
Phakopsora pachyrhizi is a biotrophic fungus, causer of the disease Asian Soybean Rust, a severe crop disease of soybean and one that demands greater investment from producers. Thus, research efforts to control this disease are still needed. We investigated the expression of metabolites in soybean plants presenting a resistant genotype inoculated with P. pachyrhizi through the untargeted metabolomics approach. The analysis was performed in control and inoculated plants with P. pachyrhizi using UHPLC-MS/MS. Principal component analysis (PCA) and the partial least squares discriminant analysis (PLS-DA), was applied to the data analysis. PCA and PLS-DA resulted in a clear separation and classification of groups between control and inoculated plants. The metabolites were putative classified and identified using the Global Natural Products Social Molecular Networking platform in flavonoids, isoflavonoids, lipids, fatty acyls, terpenes, and carboxylic acids. Flavonoids and isoflavonoids were up-regulation, while terpenes were down-regulated in response to the soybean-P. pachyrhizi interaction. Our data provide insights into the potential role of some metabolites as flavonoids and isoflavonoids in the plant resistance to ASR. This information could result in the development of resistant genotypes of soybean to P. pachyrhizi, and effective and specific products against the pathogen.
Keywords: Asian soybean rust; GNPS; Phakopsora pachyrhizi; UHPLC-MS/MS; chemometrics; metabolomics; soybean.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Unraveling Asian Soybean Rust metabolomics using mass spectrometry and Molecular Networking approach.Sci Rep. 2020 Jan 10;10(1):138. doi: 10.1038/s41598-019-56782-4. Sci Rep. 2020. PMID: 31924833 Free PMC article.
-
Soybean leaves transcriptomic data dissects the phenylpropanoid pathway genes as a defence response against Phakopsora pachyrhizi.Plant Physiol Biochem. 2018 Nov;132:424-433. doi: 10.1016/j.plaphy.2018.09.020. Epub 2018 Sep 23. Plant Physiol Biochem. 2018. PMID: 30290334
-
First Report of Asian Soybean Rust Caused by Phakopsora pachyrhizi in Puerto Rico.Plant Dis. 2013 Oct;97(10):1378. doi: 10.1094/PDIS-01-13-0108-PDN. Plant Dis. 2013. PMID: 30722174
-
Soybean-Phakopsora pachyrhizi interactions: towards the development of next-generation disease-resistant plants.Plant Biotechnol J. 2024 Feb;22(2):296-315. doi: 10.1111/pbi.14206. Epub 2023 Oct 26. Plant Biotechnol J. 2024. PMID: 37883664 Free PMC article. Review.
-
Understanding Phakopsora pachyrhizi in soybean: comprehensive insights, threats, and interventions from the Asian perspective.Front Microbiol. 2024 Jan 11;14:1304205. doi: 10.3389/fmicb.2023.1304205. eCollection 2023. Front Microbiol. 2024. PMID: 38274768 Free PMC article. Review.
Cited by
-
LC-QTOF/MS-Based Profiling of the Phytochemicals in Ice Plant (Mesembryanthemum crystallinum) and Their Bioactivities.Foods. 2024 Jun 10;13(12):1820. doi: 10.3390/foods13121820. Foods. 2024. PMID: 38928762 Free PMC article.
-
Metabolomics as a Tool to Study Underused Soy Parts: In Search of Bioactive Compounds.Foods. 2021 Jun 7;10(6):1308. doi: 10.3390/foods10061308. Foods. 2021. PMID: 34200265 Free PMC article. Review.
-
Analysis of plant metabolomics data using identification-free approaches.Appl Plant Sci. 2025 Mar 1;13(4):e70001. doi: 10.1002/aps3.70001. eCollection 2025 Jul-Aug. Appl Plant Sci. 2025. PMID: 40766901 Free PMC article. Review.
-
Phytochemical profile and anti-inflammatory activity of the hull of γ-irradiated wheat mutant lines (Triticum aestivum L.).Front Nutr. 2023 Dec 22;10:1334344. doi: 10.3389/fnut.2023.1334344. eCollection 2023. Front Nutr. 2023. PMID: 38188878 Free PMC article.
-
A comprehensive metabolome profiling of Terminalia chebula, Terminalia bellerica, and Phyllanthus emblica to explore the medicinal potential of Triphala.Sci Rep. 2024 Dec 30;14(1):31635. doi: 10.1038/s41598-024-80544-6. Sci Rep. 2024. PMID: 39738152 Free PMC article.
References
-
- Bratton M.R., Martin E.C., Elliott S., Rhodes L.V., Collins-Burow B.M., McLachlan J.A., Wiese T.E., Boue S.M., Burow M.E. Glyceollin, a novel regulator of mTOR/p70S6 in estrogen receptor positive breast cancer. J. Steroid Biochem. Mol. Biol. 2015;150:17–23. doi: 10.1016/j.jsbmb.2014.12.014. - DOI - PMC - PubMed
-
- Allen T.W., Bradley C.A., Sisson A.J., Byamukama E., Chilvers M.I., Coker C.M., Collins A.A., Damicone J.P., Dorrance A.E., Dufault N.S., et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 2017;18:19–27. doi: 10.1094/PHP-RS-16-0066. - DOI
-
- Qi M., Grayczyk J.P., Seitz J.M., Lee Y., Link T.I., Choi D., Pedley K.F., Voegele R.T., Baum T.J., Whitham S.A. Suppression or activation of immune responses by predicted secreted proteins of the soybean rust pathogen Phakopsora pachyrhizi. Mol. Plant Microbe Interact. 2018;31:163–174. doi: 10.1094/MPMI-07-17-0173-FI. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

