Transcriptome Analysis Unravels Key Factors Involved in Response to Potassium Deficiency and Feedback Regulation of K+ Uptake in Cotton Roots
- PMID: 33808570
- PMCID: PMC8003395
- DOI: 10.3390/ijms22063133
Transcriptome Analysis Unravels Key Factors Involved in Response to Potassium Deficiency and Feedback Regulation of K+ Uptake in Cotton Roots
Abstract
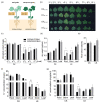
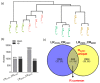
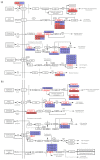
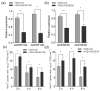
To properly understand cotton responses to potassium (K+) deficiency and how its shoot feedback regulates K+ uptake and root growth, we analyzed the changes in root transcriptome induced by low K+ (0.03 mM K+, lasting three days) in self-grafts of a K+ inefficient cotton variety (CCRI41/CCRI41, scion/rootstock) and its reciprocal grafts with a K+ efficient variety (SCRC22/CCRI41). Compared with CCRI41/CCRI41, the SCRC22 scion enhanced the K+ uptake and root growth of CCRI41 rootstock. A total of 1968 and 2539 differently expressed genes (DEGs) were identified in the roots of CCRI41/CCRI41 and SCRC22/CCRI41 in response to K+ deficiency, respectively. The overlapped and similarly (both up- or both down-) regulated DEGs in the two grafts were considered the basic response to K+ deficiency in cotton roots, whereas the DEGs only found in SCRC22/CCRI41 (1954) and those oppositely (one up- and the other down-) regulated in the two grafts might be the key factors involved in the feedback regulation of K+ uptake and root growth. The expression level of four putative K+ transporter genes (three GhHAK5s and one GhKUP3) increased in both grafts under low K+, which could enable plants to cope with K+ deficiency. In addition, two ethylene response factors (ERFs), GhERF15 and GhESE3, both down-regulated in the roots of CCRI41/CCRI41 and SCRC22/CCRI41, may negatively regulate K+ uptake in cotton roots due to higher net K+ uptake rate in their virus-induced gene silencing (VIGS) plants. In terms of feedback regulation of K+ uptake and root growth, several up-regulated DEGs related to Ca2+ binding and CIPK (CBL-interacting protein kinases), one up-regulated GhKUP3 and several up-regulated GhNRT2.1s probably play important roles. In conclusion, these results provide a deeper insight into the molecular mechanisms involved in basic response to low K+ stress in cotton roots and feedback regulation of K+ uptake, and present several low K+ tolerance-associated genes that need to be further identified and characterized.
Keywords: cotton; grafting; nutrient transporter; potassium deficiency; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

