Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions
- PMID: 33808622
- PMCID: PMC8003469
- DOI: 10.3390/nu13030992
Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions
Abstract
This work aimed to define the microbial consortia that are able to digest gluten into non-toxic and non-immunogenic peptides in the human gastrointestinal tract.
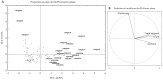
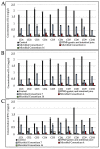
Methods: 131 out of 504 tested Bacillus and lactic acid bacteria, specifically Bacillus (64), lactobacilli (63), Pediococcus (1), and Weissella (3), showed strong gastrointestinal resistance and were selected for their PepN, PepI, PepX, PepO, and PepP activities toward synthetic substrates. Based on multivariate analysis, 24 strains were clearly distinct from the other tested strains based on having the highest enzymatic activities. As estimated by RP-HPLC and nano-ESI-MS/MS, 6 cytoplasmic extracts out of 24 selected strains showed the ability to hydrolyze immunogenic epitopes, specifically 57-68 of α9-gliadin, 62-75 of A-gliadin, 134-153 of γ-gliadin, and 57-89 (33-mer) of α2-gliadin. Live and lysed cells of selected strains were combined into different microbial consortia for hydrolyzing gluten under gastrointestinal conditions. Commercial proteolytic enzymes (Aspergillusoryzae E1, Aspergillusniger E2, Bacillussubtilis Veron HPP, and Veron PS proteases) were also added to each microbial consortium. Consortium activity was evaluated by ELISA tests, RP-HPLC-nano-ESI-MS/MS, and duodenal explants from celiac disease patients.
Results: two microbial consortia (Consortium 4: Lactiplantibacillus (Lp.) plantarum DSM33363 and DSM33364, Lacticaseibacillus (Lc.) paracasei DSM33373, Bacillussubtilis DSM33298, and Bacilluspumilus DSM33301; and Consortium 16: Lp. plantarum DSM33363 and DSM33364, Lc. paracasei DSM33373, Limosilactobacillusreuteri DSM33374, Bacillusmegaterium DSM33300, B.pumilus DSM33297 and DSM33355), containing commercial enzymes, were able to hydrolyze gluten to non-toxic and non-immunogenic peptides under gastrointestinal conditions.
Conclusions: the results of this study provide evidence that selected microbial consortia could potentially improve the digestion of gluten in gluten-sensitive patients by hydrolyzing the immunogenic peptides during gastrointestinal digestion.
Keywords: Bacillus; Lacticaseibacillus; Lactiplantibacillus; Limosilactobacillus; bacterial peptidases; gluten epitopes.
Conflict of interest statement
M.S., S.P., M.F., and B.S. declare competing interests as employees of Evonik Operations GmbH. Other authors declare no conflict of interest.
Figures


References
-
- Khan H. Genetic improvement for end-use quality in wheat. In: Qureshi A., Dar Z., Wani S., editors. Quality Breeding in Field Crops. Springer; Cham, Switzerland: 2019. pp. 239–253. - DOI
-
- Arentz–Hansen H., Mcadam S.N., Molberg Ø., Fleckenstein B., Lundin K.E., Jørgensen T.J., Jung G., Roepstorff P., Sollid L.M. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology. 2002;123:803–809. doi: 10.1053/gast.2002.35381. - DOI - PubMed
-
- Barro Losada F., Lehisa J., Giménez M.J., García-Molina M.D., Ozuna C., Comino I., Sousa Martín C., Gil Humanes J. Targeting of prolamins by RNAi in bread wheat: Effectiveness of seven silencing-fragment combinations for obtaining lines devoid of coeliac disease epitopes from highly immunogenic gliadins. Plant Biotechnol. J. 2016;14:986–996. doi: 10.1111/pbi.12455. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

