Combined Transcriptome Analysis Reveals the Ovule Abortion Regulatory Mechanisms in the Female Sterile Line of Pinus tabuliformis Carr
- PMID: 33808669
- PMCID: PMC8003466
- DOI: 10.3390/ijms22063138
Combined Transcriptome Analysis Reveals the Ovule Abortion Regulatory Mechanisms in the Female Sterile Line of Pinus tabuliformis Carr
Abstract
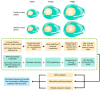
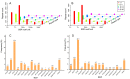
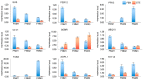
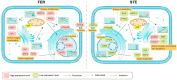
Ovule abortion is a common phenomenon in plants that has an impact on seed production. Previous studies of ovule and female gametophyte (FG) development have mainly focused on angiosperms, especially in Arabidopsis thaliana. However, because it is difficult to acquire information about ovule development in gymnosperms, this remains unclear. Here, we investigated the transcriptomic data of natural ovule abortion mutants (female sterile line, STE) and the wild type (female fertile line, FER) of Pinus tabuliformis Carr. to evaluate the mechanism of ovule abortion during the process of free nuclear mitosis (FNM). Using single-molecule real-time (SMRT) sequencing and next-generation sequencing (NGS), 18 cDNA libraries via Illumina and two normalized libraries via PacBio, with a total of almost 400,000 reads, were obtained. Our analysis showed that the numbers of isoforms and alternative splicing (AS) patterns were significantly variable between FER and STE. The functional annotation results demonstrate that genes involved in the auxin response, energy metabolism, signal transduction, cell division, and stress response were differentially expressed in different lines. In particular, AUX/IAA, ARF2, SUS, and CYCB had significantly lower expression in STE, showing that auxin might be insufficient in STE, thus hindering nuclear division and influencing metabolism. Apoptosis in STE might also have affected the expression levels of these genes. To confirm the transcriptomic analysis results, nine pairs were confirmed by quantitative real-time PCR. Taken together, these results provide new insights into ovule abortion in gymnosperms and further reveal the regulatory mechanisms of ovule development.
Keywords: Pinus tabuliformis Carr.; alternative splicing; auxin response; energy metabolism; ovule abortion; single-molecule real-time sequencing.
Conflict of interest statement
All the authors agreed on the contents of the paper and post no conflicting interest.
Figures





Similar articles
-
Screening of Differentially Expressed Genes and Localization Analysis of Female Gametophyte at the Free Nuclear Mitosis Stage in Pinus tabuliformis Carr.Int J Mol Sci. 2022 Feb 8;23(3):1915. doi: 10.3390/ijms23031915. Int J Mol Sci. 2022. PMID: 35163836 Free PMC article.
-
RNA-seq Analysis Reveals Gene Expression Profiling of Female Fertile and Sterile Ovules of PinusTabulaeformis Carr. during Free Nuclear Mitosis of the Female Gametophyte.Int J Mol Sci. 2018 Aug 1;19(8):2246. doi: 10.3390/ijms19082246. Int J Mol Sci. 2018. PMID: 30071597 Free PMC article.
-
Integrative transcriptomics and proteomics analysis constructs a new molecular model for ovule abortion in the female-sterile line of Pinus tabuliformis Carr.Plant Sci. 2020 May;294:110462. doi: 10.1016/j.plantsci.2020.110462. Epub 2020 Mar 4. Plant Sci. 2020. PMID: 32234230
-
Genome-wide transcriptome analysis of female-sterile rice ovule shed light on its abortive mechanism.Planta. 2016 Nov;244(5):1011-1028. doi: 10.1007/s00425-016-2563-x. Epub 2016 Jun 29. Planta. 2016. PMID: 27357232
-
Development and evolution of the unique ovules of flowering plants.Curr Top Dev Biol. 2019;131:373-399. doi: 10.1016/bs.ctdb.2018.10.007. Epub 2018 Dec 23. Curr Top Dev Biol. 2019. PMID: 30612624 Review.
Cited by
-
Unraveling site-specific seed formation abnormalities in Picea neoveitchii Mast. trees via widely metabolomic and transcriptomic analysis.Front Plant Sci. 2024 Dec 10;15:1495784. doi: 10.3389/fpls.2024.1495784. eCollection 2024. Front Plant Sci. 2024. PMID: 39719938 Free PMC article.
-
Development of pollinated and unpollinated ovules in Ginkgo biloba: unravelling the role of pollen in ovule tissue maturation.J Exp Bot. 2024 Jun 7;75(11):3351-3367. doi: 10.1093/jxb/erae102. J Exp Bot. 2024. PMID: 38459807 Free PMC article.
References
-
- Guo A., Zheng C. Female gametophyte development. J. Plant Biol. 2013;56:345–356. doi: 10.1007/s12374-013-0131-5. - DOI
-
- Hu Q., Gao S., Li F. Advances of research on plant female sterility. J. Beijing For. Univ. 2004;26:87–91.
-
- Rosellini D., Ferranti F., Barone P., Verones F. Expression of female sterility in alfalfa (Medicago sativa L.) Sex. Plant Reprod. 2003;15:271–279. doi: 10.1007/s00497-003-0163-y. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

