Metabolomic Variation Aligns with Two Geographically Distinct Subpopulations of Brachypodium Distachyon before and after Drought Stress
- PMID: 33808796
- PMCID: PMC8003576
- DOI: 10.3390/cells10030683
Metabolomic Variation Aligns with Two Geographically Distinct Subpopulations of Brachypodium Distachyon before and after Drought Stress
Abstract
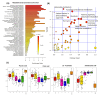
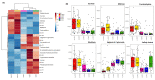
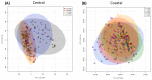
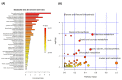
Brachypodium distachyon (Brachypodium) is a non-domesticated model grass that has been used to assess population level genomic variation. We have previously established a collection of 55 Brachypodium accessions that were sampled to reflect five different climatic regions of Turkey; designated 1a, 1c, 2, 3 and 4. Genomic and methylomic variation differentiated the collection into two subpopulations designated as coastal and central (respectively from regions 1a, 1c and the other from 2, 3 and 4) which were linked to environmental variables such as relative precipitation. Here, we assessed how far genomic variation would be reflected in the metabolomes and if this could be linked to an adaptive trait. Metabolites were extracted from eight-week-old seedlings from each accession and assessed using flow infusion high-resolution mass spectrometry (FIE-HRMS). Principal Component Analysis (PCA) of the derived metabolomes differentiated between samples from coastal and central subpopulations. The major sources of variation between seedling from the coastal and central subpopulations were identified. The central subpopulation was typified by significant increases in alanine, aspartate and glutamate metabolism and the tricarboxylic acid (TCA) cycle. Coastal subpopulation exhibited elevated levels of the auxin, indolacetic acid and rhamnose. The metabolomes of the seedling were also determined following the imposition of drought stress for seven days. The central subpopulation exhibited a metabolomic shift in response to drought, but no significant changes were seen in the coastal one. The drought responses in the central subpopulation were typified by changes in amino acids, increasing the glutamine that could be functioning as a stress signal. There were also changes in sugars that were likely to be an osmotic counter to drought, and changes in bioenergetic metabolism. These data indicate that genomic variation in our Turkish Brachypodium collection is largely reflected as distinctive metabolomes ("metabolotypes") through which drought tolerance might be mediated.
Keywords: Brachypodium distachyon; amino acids; auxin; drought; metabolome; metabolotypes; osmolytes.
Conflict of interest statement
Aleksandra Skalska is an employee of MDPI, however she is not working for the journal
Figures



References
-
- Gordon S.P., Contreras-Moreira B., Woods D.P., Des Marais D.L., Burgess D., Shu S., Stritt C., Roulin A.C., Schackwitz W., Tyler L., et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017;8:2184. doi: 10.1038/s41467-017-02292-8. - DOI - PMC - PubMed
-
- Wilson P.B., Streich J.C., Murray K.D., Eichten S.R., Cheng R., Aitken N.C., Spokas K., Warthmann N., Gordon S.P., Accession C., et al. Global diversity of the Brachypodium species complex as a resource for genome-wide association studies demonstrated for agronomic traits in response to climate. Genetics. 2019;211:317–331. doi: 10.1534/genetics.118.301589. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

