Chaperone Networks in Fungal Pathogens of Humans
- PMID: 33809191
- PMCID: PMC7998936
- DOI: 10.3390/jof7030209
Chaperone Networks in Fungal Pathogens of Humans
Abstract
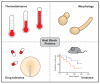
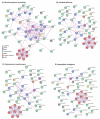
The heat shock proteins (HSPs) function as chaperones to facilitate proper folding and modification of proteins and are of particular importance when organisms are subjected to unfavourable conditions. The human fungal pathogens are subjected to such conditions within the context of infection as they are exposed to human body temperature as well as the host immune response. Herein, the roles of the major classes of HSPs are briefly reviewed and their known contributions in human fungal pathogens are described with a focus on Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus. The Hsp90s and Hsp70s in human fungal pathogens broadly contribute to thermotolerance, morphological changes required for virulence, and tolerance to antifungal drugs. There are also examples of J domain co-chaperones and small HSPs influencing the elaboration of virulence factors in human fungal pathogens. However, there are diverse members in these groups of chaperones and there is still much to be uncovered about their contributions to pathogenesis. These HSPs do not act in isolation, but rather they form a network with one another. Interactions between chaperones define their specific roles and enhance their protein folding capabilities. Recent efforts to characterize these HSP networks in human fungal pathogens have revealed that there are unique interactions relevant to these pathogens, particularly under stress conditions. The chaperone networks in the fungal pathogens are also emerging as key coordinators of pathogenesis and antifungal drug tolerance, suggesting that their disruption is a promising strategy for the development of antifungal therapy.
Keywords: antifungal drug tolerance; chaperones; fungal pathogens; heat shock proteins; thermotolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Candida albicans Heat Shock Proteins and Hsps-Associated Signaling Pathways as Potential Antifungal Targets.Front Cell Infect Microbiol. 2017 Dec 19;7:520. doi: 10.3389/fcimb.2017.00520. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29312897 Free PMC article. Review.
-
Global proteomic analyses define an environmentally contingent Hsp90 interactome and reveal chaperone-dependent regulation of stress granule proteins and the R2TP complex in a fungal pathogen.PLoS Biol. 2019 Jul 8;17(7):e3000358. doi: 10.1371/journal.pbio.3000358. eCollection 2019 Jul. PLoS Biol. 2019. PMID: 31283755 Free PMC article.
-
Dnj1 Promotes Virulence in Cryptococcus neoformans by Maintaining Robust Endoplasmic Reticulum Homeostasis Under Temperature Stress.Front Microbiol. 2021 Sep 10;12:727039. doi: 10.3389/fmicb.2021.727039. eCollection 2021. Front Microbiol. 2021. PMID: 34566931 Free PMC article.
-
Transcriptional Control of Drug Resistance, Virulence and Immune System Evasion in Pathogenic Fungi: A Cross-Species Comparison.Front Cell Infect Microbiol. 2016 Oct 20;6:131. doi: 10.3389/fcimb.2016.00131. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27812511 Free PMC article. Review.
-
The Novel J-Domain Protein Mrj1 Is Required for Mitochondrial Respiration and Virulence in Cryptococcus neoformans.mBio. 2020 Jun 9;11(3):e01127-20. doi: 10.1128/mBio.01127-20. mBio. 2020. PMID: 32518190 Free PMC article.
Cited by
-
An immunoinformatics and extensive molecular dynamics study to develop a polyvalent multi-epitope vaccine against cryptococcosis.PLoS One. 2024 Dec 31;19(12):e0315105. doi: 10.1371/journal.pone.0315105. eCollection 2024. PLoS One. 2024. Retraction in: PLoS One. 2025 Apr 4;20(4):e0322316. doi: 10.1371/journal.pone.0322316. PMID: 39739919 Free PMC article. Retracted.
-
Host-Pathogen Interaction and Resistance Mechanisms in Dermatophytes.Pathogens. 2024 Aug 4;13(8):657. doi: 10.3390/pathogens13080657. Pathogens. 2024. PMID: 39204257 Free PMC article. Review.
-
Insights and Perspectives on the Role of Proteostasis and Heat Shock Proteins in Fungal Infections.Microorganisms. 2023 Jul 25;11(8):1878. doi: 10.3390/microorganisms11081878. Microorganisms. 2023. PMID: 37630438 Free PMC article. Review.
-
Systems Biology in Fungal Research.J Fungi (Basel). 2022 May 4;8(5):478. doi: 10.3390/jof8050478. J Fungi (Basel). 2022. PMID: 35628734 Free PMC article.
-
Chaperone-Dependent Degradation of Cdc42 Promotes Cell Polarity and Shields the Protein from Aggregation.Mol Cell Biol. 2023;43(5):200-222. doi: 10.1080/10985549.2023.2198171. Epub 2023 Apr 28. Mol Cell Biol. 2023. PMID: 37114947 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

