A Novel Truncating Mutation in HOMER2 Causes Nonsyndromic Progressive DFNA68 Hearing Loss in a Spanish Family
- PMID: 33809266
- PMCID: PMC8001007
- DOI: 10.3390/genes12030411
A Novel Truncating Mutation in HOMER2 Causes Nonsyndromic Progressive DFNA68 Hearing Loss in a Spanish Family
Abstract
Nonsyndromic hereditary hearing loss is a common sensory defect in humans that is clinically and genetically highly heterogeneous. So far, 122 genes have been associated with this disorder and 50 of them have been linked to autosomal dominant (DFNA) forms like DFNA68, a rare subtype of hearing impairment caused by disruption of a stereociliary scaffolding protein (HOMER2) that is essential for normal hearing in humans and mice. In this study, we report a novel HOMER2 variant (c.832_836delCCTCA) identified in a Spanish family by using a custom NGS targeted gene panel (OTO-NGS-v2). This frameshift mutation produces a premature stop codon that may lead in the absence of NMD to a shorter variant (p.Pro278Alafs*10) that truncates HOMER2 at the CDC42 binding domain (CBD) of the coiled-coil structure, a region that is essential for protein multimerization and HOMER2-CDC42 interaction. c.832_836delCCTCA mutation is placed close to the previously identified c.840_840dup mutation found in a Chinese family that truncates the protein (p.Met281Hisfs*9) at the CBD. Functional assessment of the Chinese mutant revealed decreased protein stability, reduced ability to multimerize, and altered distribution pattern in transfected cells when compared with wild-type HOMER2. Interestingly, the Spanish and Chinese frameshift mutations might exert a similar effect at the protein level, leading to truncated mutants with the same Ct aberrant protein tail, thus suggesting that they can share a common mechanism of pathogenesis. Indeed, age-matched patients in both families display quite similar hearing loss phenotypes consisting of early-onset, moderate-to-profound progressive hearing loss. In summary, we have identified the third variant in HOMER2, which is the first one identified in the Spanish population, thus contributing to expanding the mutational spectrum of this gene in other populations, and also to clarifying the genotype-phenotype correlations of DFNA68 hearing loss.
Keywords: CDC42; HOMER2; custom panel; hereditary hearing loss; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

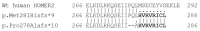
References
-
- Van Camp G., Smith R.J. Hereditary Hearing Loss Homepage. [(accessed on 20 December 2020)]; Available online: https://hereditaryhearingloss.org.
-
- Mencía A., Modamio-Høybjør S., Redshaw N., Morín M., Mayo-Merino F., Olavarrieta L., Aguirre L.A., del Castillo I., Steel K.P., Dalmay T., et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

