Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish
- PMID: 33809683
- PMCID: PMC8001686
- DOI: 10.3390/ijms22063028
Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish
Abstract
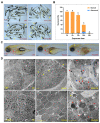
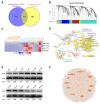
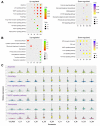
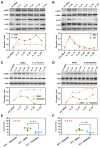
Low temperature stress represents a major threat to the lives of both farmed and wild fish species. However, biological pathways determining the development of cold resistance in fish remain largely unknown. Zebrafish larvae at 96 hpf were exposed to lethal cold stress (10 °C) for different time periods to evaluate the adverse effects at organism, tissue and cell levels. Time series RNA sequencing (RNA-seq) experiments were performed to delineate the transcriptomic landscape of zebrafish larvae under cold stress and during the subsequent rewarming phase. The genes regulated by cold stress were characterized by progressively enhanced or decreased expression, whereas the genes associated with rewarming were characterized by rapid upregulation upon return to normal temperature (28 °C). Genes such as trib3, dusp5 and otud1 were identified as the representative molecular markers of cold-induced damages through network analysis. Biological pathways involved in cold stress responses were mined from the transcriptomic data and their functions in regulating cold resistance were validated using specific inhibitors. The autophagy, FoxO and MAPK (mitogen-activated protein kinase) signaling pathways were revealed to be survival pathways for enhancing cold resistance, while apoptosis and necroptosis were the death pathways responsible for cold-induced mortality. Functional mechanisms of the survival-enhancing factors Foxo1, ERK (extracellular signal-regulated kinase) and p38 MAPK were further characterized by inhibiting their activities upon cold stress and analyzing gene expression though RNA-seq. These factors were demonstrated to determine the cold resistance of zebrafish through regulating apoptosis and p53 signaling pathway. These findings have provided novel insights into the stress responses elicited by lethal cold and shed new light on the molecular mechanisms underlying cold resistance of fish.
Keywords: biological pathways; cell death; cold resistance; stress response; transcriptional regulation; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Donaldson M.R., Cooke S.J., Patterson D.A., Macdonald J.S. Cold shock and fish. J. Fish. Biol. 2008;73:1491–1530. doi: 10.1111/j.1095-8649.2008.02061.x. - DOI
-
- Somero G.N., Hochachka P.W. Biochemical Adaptation to Environment. Am. Zool. 1971;11:159–167. doi: 10.1093/icb/11.1.159. - DOI
-
- Ou J.X., Ball J.M., Luan Y.Z., Zhao T.T., Miyagishima K.J., Xu Y.F., Zhou H.Z., Chen J.G., Merriman D.K., Xie Z., et al. iPSCs from a Hibernator Provide a Platform for Studying Cold Adaptation and Its Potential Medical Applications. Cell. 2018;173:851–863. doi: 10.1016/j.cell.2018.03.010. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2018YFD0900302-2/National Key R&D Program of China
- 31772836/National Natural Science Foundation of China
- 2021GDASYL-20210102003/Special Fund Project for Guangdong Academy of Sciences to Build Domestic First-class Research institutions
- 2016305/Youth Innovation Promotion Association of Chinese Academy of Sciences
- 2020B0202080005/Key-Area Research and Development Program of Guangdong Province
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

