Characterisation of a G2P[4] Rotavirus Outbreak in Western Australia, Predominantly Impacting Aboriginal Children
- PMID: 33809709
- PMCID: PMC8002226
- DOI: 10.3390/pathogens10030350
Characterisation of a G2P[4] Rotavirus Outbreak in Western Australia, Predominantly Impacting Aboriginal Children
Abstract
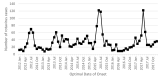
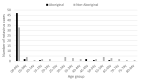
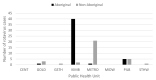
In May, 2017, an outbreak of rotavirus gastroenteritis was reported that predominantly impacted Aboriginal children ≤4 years of age in the Kimberley region of Western Australia. G2P[4] was identified as the dominant genotype circulating during this period and polyacrylamide gel electrophoresis revealed the majority of samples exhibited a conserved electropherotype. Full genome sequencing was performed on representative samples that exhibited the archetypal DS-1-like genome constellation: G2-P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2 and phylogenetic analysis revealed all genes of the outbreak samples were closely related to contemporary Japanese G2P[4] samples. The outbreak samples consistently fell within conserved sub-clades comprised of Hungarian and Australian G2P[4] samples from 2010. The 2017 outbreak variant was not closely related to G2P[4] variants associated with prior outbreaks in Aboriginal communities in the Northern Territory. When compared to the G2 component of the RotaTeq vaccine, the outbreak variant exhibited mutations in known antigenic regions; however, these mutations are frequently observed in contemporary G2P[4] strains. Despite the level of vaccine coverage achieved in Australia, outbreaks continue to occur in vaccinated populations, which pose challenges to regional areas and remote communities. Continued surveillance and characterisation of emerging variants are imperative to ensure the ongoing success of the rotavirus vaccination program in Australia.
Keywords: Aboriginal; G2P[4]; Indigenous; Western Australia; gastroenteritis; outbreak; rotavirus; vaccine; whole genome sequencing.
Conflict of interest statement
The Australian Rotavirus Surveillance Program is supported by research grants from the vaccine companies Commonwealth Serum Laboratories (bioCSL)/Sequis (2010–2018) and GlaxoSmithKline (2010–2016, study ID116120 2017–2018). C.M.D. has served on an advisory board for GSK (2019), all payments were paid directly to an administrative fund held by Murdoch Children’s Research Institute. All other authors declare no competing interests.
Figures






References
-
- Troeger C., Khalil I.A., Rao P.C., Cao S., Blacker B.F., Ahmed T., Armah G., Bines J.E., Brewer T.G., Colombara D.V., et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018;172:958–965. doi: 10.1001/jamapediatrics.2018.1960. - DOI - PMC - PubMed
-
- World Health Organization Vaccine in National Immunization Programme Update. [(accessed on 5 May 2020)]; Available online: www.who.int/immunization/monitoring_surveillance/VaccineIntroStatus.pptx....
-
- Roczo-Farkas S., Kirkwood C.D., Cowley D., Barnes G.L., Bishop R.F., Bogdanovic-Sakran N., Boniface K., Donato C.M., Bines J.E. The Impact of Rotavirus Vaccines on Genotype Diversity: A Comprehensive Analysis of 2 Decades of Australian Surveillance Data. J. Infect. Dis. 2018;218:546–554. doi: 10.1093/infdis/jiy197. - DOI - PubMed
-
- Rotavirus Classification Working Group List of Accepted Genotypes. [(accessed on 18 January 2021)]; Available online: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

