Design and Evaluation of a Macroarray for Detection, Identification, and Typing of Viral Hemorrhagic Septicemia Virus (VHSV)
- PMID: 33809757
- PMCID: PMC8002285
- DOI: 10.3390/ani11030841
Design and Evaluation of a Macroarray for Detection, Identification, and Typing of Viral Hemorrhagic Septicemia Virus (VHSV)
Abstract
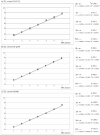
The viral hemorrhagic septicemia virus (VHSV) is the causative agent of an important disease in freshwater and marine fishes. Its diagnosis officially relies on the isolation of the virus in cell culture and its identification by serological or polymerase chain reaction (PCR) methodologies. Nowadays, reverse transcription real-time quantitative PCR (RT-qPCR) is the most widely employed technique for the detection of this virus and some studies have reported the validation of RT-qPCR procedures for the detection, typing, and quantification of VHSV isolates. However, although the efficacy of this technique is not in doubt, it can be cumbersome and even impractical when it comes to processing large numbers of samples, a situation in which cross-contamination problems cannot be ruled out. In the present study, we have designed and validated a macroarray for the simultaneous detection, typing, and quantification of VHSV strains. Its analytical sensitivity (5-50 TCID50/mL), analytical specificity (intra and intergroup), efficiency (E = 100.0-101.1) and reliability (repeatability and reproducibility with CV < 5%, and standard curves with R2 < 0.95) with strains from any VHSV genotype have been widely demonstrated. The procedure is based on the 'binary multiplex RT-qPCR system (bmRT-qPCR)' previously reported by the same team, applied to arrays of 96-well PCR strip tubes plates, which can be stored at -25 °C for three months and up to one year before their use, without significant loss of efficiency.
Keywords: diagnosis; viral haemorrhagic septicaemia virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Smail D.A. Viral haemorrhagic septicaemia. In: Woo P.T.K., Bruno D.W., editors. Fish Diseases and Disorders, Volume 3: Viral, Bacterial and Fungal Infections. CABI Publishing; New York, NY, USA: 1999. pp. 123–147.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

