Deep Learning for Novel Antimicrobial Peptide Design
- PMID: 33810011
- PMCID: PMC8004669
- DOI: 10.3390/biom11030471
Deep Learning for Novel Antimicrobial Peptide Design
Abstract
Antimicrobial resistance is an increasing issue in healthcare as the overuse of antibacterial agents rises during the COVID-19 pandemic. The need for new antibiotics is high, while the arsenal of available agents is decreasing, especially for the treatment of infections by Gram-negative bacteria like Escherichia coli. Antimicrobial peptides (AMPs) are offering a promising route for novel antibiotic development and deep learning techniques can be utilised for successful AMP design. In this study, a long short-term memory (LSTM) generative model and a bidirectional LSTM classification model were constructed to design short novel AMP sequences with potential antibacterial activity against E. coli. Two versions of the generative model and six versions of the classification model were trained and optimised using Bayesian hyperparameter optimisation. These models were used to generate sets of short novel sequences that were classified as antimicrobial or non-antimicrobial. The validation accuracies of the classification models were 81.6-88.9% and the novel AMPs were classified as antimicrobial with accuracies of 70.6-91.7%. Predicted three-dimensional conformations of selected short AMPs exhibited the alpha-helical structure with amphipathic surfaces. This demonstrates that LSTMs are effective tools for generating novel AMPs against targeted bacteria and could be utilised in the search for new antibiotics leads.
Keywords: Escherichia coli; antimicrobial peptides; deep learning; long short-term memory; machine learning; peptide design.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
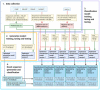
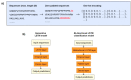
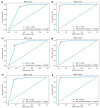
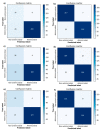

Similar articles
-
Rational design of antimicrobial peptides targeting Gram-negative bacteria.Comput Biol Chem. 2021 Jun;92:107475. doi: 10.1016/j.compbiolchem.2021.107475. Epub 2021 Mar 17. Comput Biol Chem. 2021. PMID: 33813188
-
Design, Engineering and Discovery of Novel α-Helical and β-Boomerang Antimicrobial Peptides against Drug Resistant Bacteria.Int J Mol Sci. 2020 Aug 11;21(16):5773. doi: 10.3390/ijms21165773. Int J Mol Sci. 2020. PMID: 32796755 Free PMC article. Review.
-
An Smp43-Derived Short-Chain α-Helical Peptide Displays a Unique Sequence and Possesses Antimicrobial Activity against Both Gram-Positive and Gram-Negative Bacteria.Toxins (Basel). 2021 May 11;13(5):343. doi: 10.3390/toxins13050343. Toxins (Basel). 2021. PMID: 34064808 Free PMC article.
-
Design of new truncated derivatives based on direct and reverse mirror repeats of first six residues of Caerin 4 antimicrobial peptide and evaluation of their activity and cytotoxicity.Chem Biol Drug Des. 2020 Aug;96(2):801-811. doi: 10.1111/cbdd.13689. Epub 2020 Apr 19. Chem Biol Drug Des. 2020. PMID: 32259385
-
Chemically modified and conjugated antimicrobial peptides against superbugs.Chem Soc Rev. 2021 Apr 26;50(8):4932-4973. doi: 10.1039/d0cs01026j. Chem Soc Rev. 2021. PMID: 33710195 Review.
Cited by
-
AMPlify: attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens.BMC Genomics. 2022 Jan 25;23(1):77. doi: 10.1186/s12864-022-08310-4. BMC Genomics. 2022. PMID: 35078402 Free PMC article.
-
Discovery of Lactomodulin, a Unique Microbiome-Derived Peptide That Exhibits Dual Anti-Inflammatory and Antimicrobial Activity against Multidrug-Resistant Pathogens.Int J Mol Sci. 2023 Apr 7;24(8):6901. doi: 10.3390/ijms24086901. Int J Mol Sci. 2023. PMID: 37108065 Free PMC article.
-
DRAMP 4.0: an open-access data repository dedicated to the clinical translation of antimicrobial peptides.Nucleic Acids Res. 2025 Jan 6;53(D1):D403-D410. doi: 10.1093/nar/gkae1046. Nucleic Acids Res. 2025. PMID: 39526377 Free PMC article.
-
Harnessing of Artificial Intelligence for the Diagnosis and Prevention of Hospital-Acquired Infections: A Systematic Review.Diagnostics (Basel). 2024 Feb 23;14(5):484. doi: 10.3390/diagnostics14050484. Diagnostics (Basel). 2024. PMID: 38472956 Free PMC article. Review.
-
Tackling the Antimicrobial Resistance "Pandemic" with Machine Learning Tools: A Summary of Available Evidence.Microorganisms. 2024 Apr 23;12(5):842. doi: 10.3390/microorganisms12050842. Microorganisms. 2024. PMID: 38792673 Free PMC article. Review.
References
-
- Petrosillo N. Infections: The Emergency of the New Millennium. In: Signore A., Glaudemans A.W.J.M., editors. Nuclear Medicine in Infectious Diseases. Springer; Berlin/Heidelberg, Germany: 2019. pp. 1–8.
-
- O’Neil J. Review on Antimicrobial Resistance. AMR-Review; London, UK: 2016. Tackling drug-resistant infections globally: Final report and recommendations; pp. 1–84.
-
- International Severe Acute Respiratory and Emerging Infection Consortium COVID-19 Report. [(accessed on 10 June 2020)];2020 Available online: https://media.tghn.org/medialibrary/2020/04/ISARIC_Data_Platform_COVID-1....
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

