Differential MicroRNA Expression Involved in Endometrial Receptivity of Goats
- PMID: 33810054
- PMCID: PMC8004627
- DOI: 10.3390/biom11030472
Differential MicroRNA Expression Involved in Endometrial Receptivity of Goats
Abstract
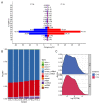
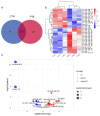
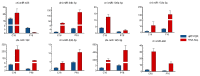
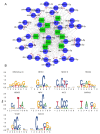
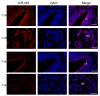
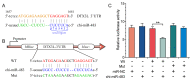
Endometrial receptivity represents one of the leading factors affecting the successful implantation of embryos during early pregnancy. However, the mechanism of microRNAs (miRNAs) to establish goat endometrial receptivity remains unclear. This study was intended to identify potential miRNAs and regulatory mechanisms associated with establishing endometrial receptivity through integrating bioinformatics analysis and experimental verification. MiRNA expression profiles were obtained by high-throughput sequencing, resulting in the detection of 33 differentially expressed miRNAs (DEMs), followed by their validation through quantitative RT-PCR. Furthermore, 10 potential transcription factors (TFs) and 1316 target genes of these DEMs were obtained, and the TF-miRNA and miRNA-mRNA interaction networks were constructed. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses indicated that these miRNAs were significantly linked to establishing endometrial receptivity. Moreover, the fluorescence in situ hybridization (FISH) analysis, dual-luciferase report assay, and immunohistochemistry (IHC) analysis corroborated that chi-miR-483 could directly bind to deltex E3 ubiquitin ligase 3L (DTX3L) to reduce its expression level. In conclusion, our findings contribute to a better understanding of molecular mechanisms regulating the endometrial receptivity of goats, and they provide a reference for improving embryo implantation efficiency.
Keywords: DTX3L; chi-miR-483; endometrium receptivity; goat; high-throughput sequencing; implantation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Integrating Analysis to Identify Differential circRNAs Involved in Goat Endometrial Receptivity.Int J Mol Sci. 2023 Jan 12;24(2):1531. doi: 10.3390/ijms24021531. Int J Mol Sci. 2023. PMID: 36675045 Free PMC article.
-
Differential Expression Pattern of Goat Uterine Fluids Extracellular Vesicles miRNAs during Peri-Implantation.Cells. 2021 Sep 3;10(9):2308. doi: 10.3390/cells10092308. Cells. 2021. PMID: 34571957 Free PMC article.
-
[Differential expression of microRNA in eutopic endometrium tissue during implantation window for patients with endometriosis related infertility].Zhonghua Fu Chan Ke Za Zhi. 2016 Jun 25;51(6):436-41. doi: 10.3760/cma.j.issn.0529-567X.2016.06.007. Zhonghua Fu Chan Ke Za Zhi. 2016. PMID: 27356479 Chinese.
-
The role of microRNAs in the regulation of critical genes and signalling pathways that determine endometrial receptivity.Zygote. 2024 Aug;32(4):261-270. doi: 10.1017/S0967199424000297. Epub 2024 Sep 18. Zygote. 2024. PMID: 39291681 Review.
-
MicroRNAs in the Regulation of Endometrial Receptivity for Embryo Implantation.Int J Mol Sci. 2022 Jun 1;23(11):6210. doi: 10.3390/ijms23116210. Int J Mol Sci. 2022. PMID: 35682889 Free PMC article. Review.
Cited by
-
miRNA expression profiles of peripheral white blood cells from beef heifers with varying reproductive potential.Front Genet. 2023 May 10;14:1174145. doi: 10.3389/fgene.2023.1174145. eCollection 2023. Front Genet. 2023. PMID: 37234872 Free PMC article.
-
Differential MicroRNA Expression in Porcine Endometrium Related to Spontaneous Embryo Loss during Early Pregnancy.Int J Mol Sci. 2022 Jul 24;23(15):8157. doi: 10.3390/ijms23158157. Int J Mol Sci. 2022. PMID: 35897733 Free PMC article.
-
Bioinformatic analysis of endometrial miRNA expression profile at day 26-28 of pregnancy in the mare.Sci Rep. 2024 Feb 16;14(1):3900. doi: 10.1038/s41598-024-53499-x. Sci Rep. 2024. PMID: 38365979 Free PMC article.
-
Circulating miRNAs in maternal plasma as potential biomarkers of early pregnancy in sheep.Front Genet. 2022 Aug 17;13:929477. doi: 10.3389/fgene.2022.929477. eCollection 2022. Front Genet. 2022. PMID: 36061213 Free PMC article.
-
Integrating Analysis to Identify Differential circRNAs Involved in Goat Endometrial Receptivity.Int J Mol Sci. 2023 Jan 12;24(2):1531. doi: 10.3390/ijms24021531. Int J Mol Sci. 2023. PMID: 36675045 Free PMC article.
References
-
- Geisert R., Schmitt R. Early embryonic survival in the pig: Can it be improved? J. Anim. Sci. 2002;80:E54–E65.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

