Multiple Early Introductions of SARS-CoV-2 to Cape Town, South Africa
- PMID: 33810168
- PMCID: PMC8005015
- DOI: 10.3390/v13030526
Multiple Early Introductions of SARS-CoV-2 to Cape Town, South Africa
Abstract
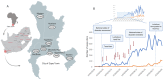
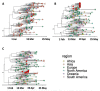
Cape Town was the first city in South Africa to experience the full impact of the coronavirus disease 2019 (COVID-19) pandemic. We acquired samples from all suspected cases and their contacts during the first month of the pandemic from Tygerberg Hospital. Nanopore sequencing generated SARS-CoV-2 whole genomes. Phylogenetic inference with maximum likelihood and Bayesian methods were used to determine lineages that seeded the local epidemic. Three patients were known to have travelled internationally and an outbreak was detected in a nearby supermarket. Sequencing of 50 samples produced 46 high-quality genomes. The sequences were classified as lineages: B, B.1, B.1.1.1, B.1.1.161, B.1.1.29, B.1.8, B.39, and B.40. All the sequences from persons under investigation (PUIs) in the supermarket outbreak (lineage B.1.8) fall within a clade from the Netherlands with good support (p > 0.9). In addition, a new mutation, 5209A>G, emerged within the Cape Town cluster. The molecular clock analysis suggests that this occurred around 13 March 2020 (95% confidence interval: 9-17 March). The phylogenetic reconstruction suggests at least nine early introductions of SARS-CoV-2 into Cape Town and an early localized transmission in a shopping environment. Genomic surveillance was successfully used to investigate and track the spread of early introductions of SARS-CoV-2 in Cape Town.
Keywords: COVID-19; Cape Town; SARS-CoV-2; South Africa; Western Cape Province; betacoronavirus; genome sequencing; molecular epidemiology; mutation; phylogenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
-
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J., Gubareva L.V., Xu X., Bridges C.B., et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. Erratum in 2009, 361, 102. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

