Breast Cancer Patient Prognosis Is Determined by the Interplay between TP53 Mutation and Alternative Transcript Expression: Insights from TP53 Long Amplicon Digital PCR Assays
- PMID: 33810361
- PMCID: PMC8036703
- DOI: 10.3390/cancers13071531
Breast Cancer Patient Prognosis Is Determined by the Interplay between TP53 Mutation and Alternative Transcript Expression: Insights from TP53 Long Amplicon Digital PCR Assays
Abstract
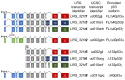
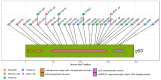
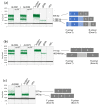
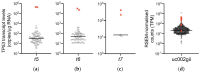
The TP53 gene locus is capable of producing multiple RNA transcripts encoding the different p53 protein isoforms. We recently described multiplex long amplicon droplet digital PCR (ddPCR) assays to quantify seven of eight TP53 reference transcripts in human tumors. Here, we describe a new long amplicon ddPCR assay to quantify expression of the eighth TP53 reference transcript encoding ∆40p53α. We then applied these assays, alongside DNA sequencing of the TP53 gene locus, to tumors from a cohort of New Zealand (NZ) breast cancer patients. We found a high prevalence of mutations at TP53 splice sites in the NZ breast cancer cohort. Mutations at TP53 intron 4 splice sites were associated with overexpression of ∆133TP53 transcripts. Cox proportional hazards survival analysis showed that interplay between TP53 mutation status and expression of TP53 transcript variants was significantly associated with patient outcome, over and above standard clinical and pathological information. In particular, patients with no TP53 mutation and a low ratio of TP53 transcripts t2 to t1, which derive from alternative intron 1 acceptor splice sites, had a remarkably good outcome. We suggest that this type of analysis, integrating mutation and transcript expression, provides a step-change in our understanding of TP53 in cancer.
Keywords: TP53 isoforms; alternative splicing; breast cancer prognosis; long amplicon digital PCR.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Pereira B., Chin S.-F., Rueda O.M., Vollan H.-K.M., Provenzano E., Bardwell H.A., Pugh M., Jones L.A., Russell R., Sammut S.-J., et al. The somatic mutation profiles of 2433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

