Tracing dsDNA Virus-Host Coevolution through Correlation of Their G-Quadruplex-Forming Sequences
- PMID: 33810462
- PMCID: PMC8036883
- DOI: 10.3390/ijms22073433
Tracing dsDNA Virus-Host Coevolution through Correlation of Their G-Quadruplex-Forming Sequences
Abstract
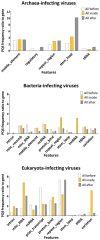
The importance of gene expression regulation in viruses based upon G-quadruplex may point to its potential utilization in therapeutic targeting. Here, we present analyses as to the occurrence of putative G-quadruplex-forming sequences (PQS) in all reference viral dsDNA genomes and evaluate their dependence on PQS occurrence in host organisms using the G4Hunter tool. PQS frequencies differ across host taxa without regard to GC content. The overlay of PQS with annotated regions reveals the localization of PQS in specific regions. While abundance in some, such as repeat regions, is shared by all groups, others are unique. There is abundance within introns of Eukaryota-infecting viruses, but depletion of PQS in introns of bacteria-infecting viruses. We reveal a significant positive correlation between PQS frequencies in dsDNA viruses and corresponding hosts from archaea, bacteria, and eukaryotes. A strong relationship between PQS in a virus and its host indicates their close coevolution and evolutionarily reciprocal mimicking of genome organization.
Keywords: G-quadruplex; G4Hunter; bioinformatics; coevolution; dsDNA; host; virus.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Analyses of viral genomes for G-quadruplex forming sequences reveal their correlation with the type of infection.Biochimie. 2021 Jul;186:13-27. doi: 10.1016/j.biochi.2021.03.017. Epub 2021 Apr 9. Biochimie. 2021. PMID: 33839192
-
G-quadruplexes in genomes of viruses infecting eukaryotes or prokaryotes are under different selection pressures from hosts.J Genet Genomics. 2022 Jan;49(1):20-29. doi: 10.1016/j.jgg.2021.08.018. Epub 2021 Sep 30. J Genet Genomics. 2022. PMID: 34601118
-
G-quadruplexes in H1N1 influenza genomes.BMC Genomics. 2021 Jan 23;22(1):77. doi: 10.1186/s12864-021-07377-9. BMC Genomics. 2021. PMID: 33485319 Free PMC article.
-
The LUCA and its complex virome.Nat Rev Microbiol. 2020 Nov;18(11):661-670. doi: 10.1038/s41579-020-0408-x. Epub 2020 Jul 14. Nat Rev Microbiol. 2020. PMID: 32665595 Review.
-
Above and Beyond Watson and Crick: Guanine Quadruplex Structures and Microbes.Annu Rev Microbiol. 2018 Sep 8;72:49-69. doi: 10.1146/annurev-micro-090817-062629. Epub 2018 May 31. Annu Rev Microbiol. 2018. PMID: 29852085 Review.
Cited by
-
Contrasting roles for G-quadruplexes in regulating human Bcl-2 and virus homologues KSHV KS-Bcl-2 and EBV BHRF1.Sci Rep. 2022 Mar 23;12(1):5019. doi: 10.1038/s41598-022-08161-9. Sci Rep. 2022. PMID: 35322051 Free PMC article.
-
G-quadruplex occurrence and conservation: more than just a question of guanine-cytosine content.NAR Genom Bioinform. 2022 Mar 4;4(1):lqac010. doi: 10.1093/nargab/lqac010. eCollection 2022 Mar. NAR Genom Bioinform. 2022. PMID: 35261973 Free PMC article.
-
G-quadruplexes in the evolution of hepatitis B virus.Nucleic Acids Res. 2023 Aug 11;51(14):7198-7204. doi: 10.1093/nar/gkad556. Nucleic Acids Res. 2023. PMID: 37395407 Free PMC article.
-
Impacts of Molecular Structure on Nucleic Acid-Protein Interactions.Int J Mol Sci. 2022 Dec 26;24(1):407. doi: 10.3390/ijms24010407. Int J Mol Sci. 2022. PMID: 36613851 Free PMC article.
-
G-quadruplexes in helminth parasites.Nucleic Acids Res. 2022 Mar 21;50(5):2719-2735. doi: 10.1093/nar/gkac129. Nucleic Acids Res. 2022. PMID: 35234933 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

