Persistent Southern Tomato Virus (STV) Interacts with Cucumber Mosaic and/or Pepino Mosaic Virus in Mixed- Infections Modifying Plant Symptoms, Viral Titer and Small RNA Accumulation
- PMID: 33810543
- PMCID: PMC8066132
- DOI: 10.3390/microorganisms9040689
Persistent Southern Tomato Virus (STV) Interacts with Cucumber Mosaic and/or Pepino Mosaic Virus in Mixed- Infections Modifying Plant Symptoms, Viral Titer and Small RNA Accumulation
Abstract
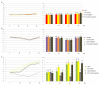
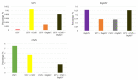
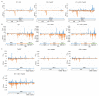
Southern tomato virus (STV) is a persistent virus that was, at the beginning, associated with some tomato fruit disorders. Subsequent studies showed that the virus did not induce apparent symptoms in single infections. Accordingly, the reported symptoms could be induced by the interaction of STV with other viruses, which frequently infect tomato. Here, we studied the effect of STV in co- and triple-infections with Cucumber mosaic virus (CMV) and Pepino mosaic virus (PepMV). Our results showed complex interactions among these viruses. Co-infections leaded to a synergism between STV and CMV or PepMV: STV increased CMV titer and plant symptoms at early infection stages, whereas PepMV only exacerbated the plant symptoms. CMV and PepMV co-infection showed an antagonistic interaction with a strong decrease of CMV titer and a modification of the plant symptoms with respect to the single infections. However, the presence of STV in a triple-infection abolished this antagonism, restoring the CMV titer and plant symptoms. The siRNAs analysis showed a total of 78 miRNAs, with 47 corresponding to novel miRNAs in tomato, which were expressed differentially in the plants that were infected with these viruses with respect to the control mock-inoculated plants. These miRNAs were involved in the regulation of important functions and their number and expression level varied, depending on the virus combination. The number of vsiRNAs in STV single-infected tomato plants was very small, but STV vsiRNAs increased with the presence of CMV and PepMV. Additionally, the rates of CMV and PepMV vsiRNAs varied depending on the virus combination. The frequencies of vsiRNAs in the viral genomes were not uniform, but they were not influenced by other viruses.
Keywords: Amalgaviridae; antagonism; miRNAs; mixed-infections; persistent virus; synergism; vsiRNAs.
Conflict of interest statement
The authors declare that they have no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
First Report of Pepino mosaic virus Infecting Greenhouse Cherry Tomatoes in Greece.Plant Dis. 2011 Jan;95(1):78. doi: 10.1094/PDIS-09-10-0643. Plant Dis. 2011. PMID: 30743682
-
Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants.Viruses. 2020 Aug 11;12(8):879. doi: 10.3390/v12080879. Viruses. 2020. PMID: 32796777 Free PMC article.
-
First Report of Tomato torrado virus Infecting Tomato in Single and Mixed Infections with Cucumber mosaic virus in Panama.Plant Dis. 2009 Feb;93(2):198. doi: 10.1094/PDIS-93-2-0198A. Plant Dis. 2009. PMID: 30764131
-
Natural Occurrence of Viruses in Lycopersicon spp. in Ecuador.Plant Dis. 2005 Nov;89(11):1244. doi: 10.1094/PD-89-1244C. Plant Dis. 2005. PMID: 30786461
-
Pepino mosaic virus: a successful pathogen that rapidly evolved from emerging to endemic in tomato crops.Mol Plant Pathol. 2010 Mar;11(2):179-89. doi: 10.1111/j.1364-3703.2009.00600.x. Mol Plant Pathol. 2010. PMID: 20447268 Free PMC article. Review.
Cited by
-
Translating virome analyses to support biosecurity, on-farm management, and crop breeding.Front Plant Sci. 2023 Mar 14;14:1056603. doi: 10.3389/fpls.2023.1056603. eCollection 2023. Front Plant Sci. 2023. PMID: 36998684 Free PMC article. Review.
-
High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family.Plants (Basel). 2022 Jun 23;11(13):1665. doi: 10.3390/plants11131665. Plants (Basel). 2022. PMID: 35807616 Free PMC article.
-
N Protein of Tomato Spotted Wilt Virus Proven to Be Antagonistic Against Tomato Yellow Leaf Curl Virus in Nicotiana benthamiana.Mol Plant Pathol. 2025 Jan;26(1):e70046. doi: 10.1111/mpp.70046. Mol Plant Pathol. 2025. PMID: 39740810 Free PMC article.
-
Molecular Detection of Southern Tomato Amalgavirus Prevalent in Tomatoes and Its Genomic Characterization with Global Evolutionary Dynamics.Viruses. 2022 Nov 9;14(11):2481. doi: 10.3390/v14112481. Viruses. 2022. PMID: 36366579 Free PMC article.
-
The Potential of Molecular Indicators of Plant Virus Infection: Are Plants Able to Tell Us They Are Infected?Plants (Basel). 2022 Jan 11;11(2):188. doi: 10.3390/plants11020188. Plants (Basel). 2022. PMID: 35050076 Free PMC article.
References
-
- Souiri A., Khataby K., Kasmi Y., Zemzami M., Amzazi S., Ennaji M.M. Emerging and Reemerging Viral Pathogens: Volume 2: Applied Virology Approaches Related to Human, Animal and Environmental Pathogens. Elsevier; Amsterdam, The Netherlands: 2019. Risk assessment and biosecurity considerations in control of emergent plant viruses; pp. 287–311.
-
- Elvira-González L., Carpino C., Alfaro-Fernández A., Font-San Ambrosio M.I., Peiró R., Rubio L., Galipienso L. A sensitive real-time RT-PCR reveals a high incidence of Southern tomato virus (STV) in Spanish tomato crops. Spanish J. Agric. Res. 2018;16:1008. doi: 10.5424/sjar/2018163-12961. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

