Transcriptomic Analyses Shed Light on Critical Genes Associated with Bibenzyl Biosynthesis in Dendrobium officinale
- PMID: 33810588
- PMCID: PMC8065740
- DOI: 10.3390/plants10040633
Transcriptomic Analyses Shed Light on Critical Genes Associated with Bibenzyl Biosynthesis in Dendrobium officinale
Abstract
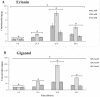
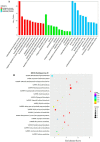
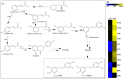
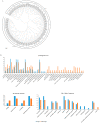
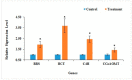
The Dendrobium plants (members of the Orchidaceae family) are used as traditional Chinese medicinal herbs. Bibenzyl, one of the active compounds in Dendrobium officinale, occurs in low amounts among different tissues. However, market demands require a higher content of thes compounds to meet the threshold for drug production. There is, therefore, an immediate need to dissect the physiological and molecular mechanisms underlying how bibenzyl compounds are biosynthesized in D. officinale tissues. In this study, the accumulation of erianin and gigantol in tissues were studied as representative compounds of bibenzyl. Exogenous application of Methyl-Jasmonate (MeJA) promotes the biosynthesis of bibenzyl compounds; therefore, transcriptomic analyses were conducted between D. officinale-treated root tissues and a control. Our results show that the root tissues contained the highest content of bibenzyl (erianin and gigantol). We identified 1342 differentially expressed genes (DEGs) with 912 up-regulated and 430 down-regulated genes in our transcriptome dataset. Most of the identified DEGs are functionally involved in the JA signaling pathway and the biosynthesis of secondary metabolites. We also identified two candidate cytochrome P450 genes and nine other enzymatic genes functionally involved in bibenzyl biosynthesis. Our study provides insights on the identification of critical genes associated with bibenzyl biosynthesis and accumulation in Dendrobium plants, paving the way for future research on dissecting the physiological and molecular mechanisms of bibenzyl synthesis in plants as well as guide genetic engineering for the improvement of Dendrobium varieties through increasing bibenzyl content for drug production and industrialization.
Keywords: Dendrobium officinale; bibenzyl; erianin; gigantol; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Research Advances in Multi-Omics on the Traditional Chinese Herb Dendrobium officinale.Front Plant Sci. 2022 Jan 11;12:808228. doi: 10.3389/fpls.2021.808228. eCollection 2021. Front Plant Sci. 2022. PMID: 35087561 Free PMC article. Review.
-
Integrated metabolomic and transcriptomic analyses of Dendrobium chrysotoxum and D. thyrsiflorum reveal the biosynthetic pathway from gigantol to erianin.Front Plant Sci. 2024 Sep 26;15:1436560. doi: 10.3389/fpls.2024.1436560. eCollection 2024. Front Plant Sci. 2024. PMID: 39391777 Free PMC article.
-
Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale.J Plant Res. 2019 May;132(3):419-429. doi: 10.1007/s10265-019-01099-6. Epub 2019 Mar 22. J Plant Res. 2019. PMID: 30903398
-
Functional Insights into the Roles of Hormones in the Dendrobium officinale-Tulasnella sp. Germinated Seed Symbiotic Association.Int J Mol Sci. 2018 Nov 6;19(11):3484. doi: 10.3390/ijms19113484. Int J Mol Sci. 2018. PMID: 30404159 Free PMC article.
-
Transcriptomic Landscape of Medicinal Dendrobium Reveals Genes Associated With the Biosynthesis of Bioactive Components.Front Plant Sci. 2020 Apr 28;11:391. doi: 10.3389/fpls.2020.00391. eCollection 2020. Front Plant Sci. 2020. PMID: 32411153 Free PMC article. Review.
Cited by
-
A chromosome-level Dendrobium moniliforme genome assembly reveals the regulatory mechanisms of flavonoid and carotenoid biosynthesis pathways.Acta Pharm Sin B. 2025 Apr;15(4):2253-2272. doi: 10.1016/j.apsb.2025.03.005. Epub 2025 Mar 7. Acta Pharm Sin B. 2025. PMID: 40486862 Free PMC article.
-
Genome-wide identification and characterization of active ingredients related β-Glucosidases in Dendrobium catenatum.BMC Genomics. 2022 Aug 23;23(1):612. doi: 10.1186/s12864-022-08840-x. BMC Genomics. 2022. PMID: 35999493 Free PMC article.
-
Research Advances in Multi-Omics on the Traditional Chinese Herb Dendrobium officinale.Front Plant Sci. 2022 Jan 11;12:808228. doi: 10.3389/fpls.2021.808228. eCollection 2021. Front Plant Sci. 2022. PMID: 35087561 Free PMC article. Review.
-
Integrated metabolomic and transcriptomic analyses of Dendrobium chrysotoxum and D. thyrsiflorum reveal the biosynthetic pathway from gigantol to erianin.Front Plant Sci. 2024 Sep 26;15:1436560. doi: 10.3389/fpls.2024.1436560. eCollection 2024. Front Plant Sci. 2024. PMID: 39391777 Free PMC article.
-
The Transcriptome Profiling of Flavonoids and Bibenzyls Reveals Medicinal Importance of Rare Orchid Arundina graminifolia.Front Plant Sci. 2022 Jun 23;13:923000. doi: 10.3389/fpls.2022.923000. eCollection 2022. Front Plant Sci. 2022. PMID: 35812923 Free PMC article.
References
-
- Goossens A.H., Laakso S.T., Seppanen-Laakso I., Biondi T., De Sutter S., Lammertyn V., Nuutila F., Soderlund A.M., Zabeau H., Inze M., et al. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. USA. 2003;100:8595–8600. doi: 10.1073/pnas.1032967100. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

